Development of a PCR-restriction fragment length polymorphism assay for the epidemiological analysis of Shiga toxin-producing Escherichia coli
- PMID: 15528716
- PMCID: PMC525232
- DOI: 10.1128/JCM.42.11.5205-5213.2004
Development of a PCR-restriction fragment length polymorphism assay for the epidemiological analysis of Shiga toxin-producing Escherichia coli
Abstract
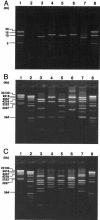
Six characteristic regions (I to VI) were identified in Shiga toxin 2 (Stx2) phages (T. Sato, T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki, Gene 309:35-48, 2003). Region V, which is ca. 10 kb in size and is located in the upstream region of the Stx operons, includes the most distinctive region among six Stx phages whose genome sequences have been determined. In this study, we developed a PCR-restriction fragment length polymorphism (RFLP) assay for the epidemiological analysis of Shiga toxin-producing Escherichia coli (STEC) on the basis of the diversity of region V. When region V was amplified by long and accurate-PCR (LA-PCR) with five control E. coli strains carrying six different Stx phages such as E. coli strains C600 (Stx1 phage), C600 (933W phage), C600 (Stx2 phage-I), C600 (Stx2 phage-II), and O157:H7 Sakai strain RIMD0509952 (VT1-Sakai phage and VT2-Sakai phage), an expected size of the band was obtained. Restriction digest of each PCR product with BglI or EcoRV also gave the expected sizes of banding patterns and discriminated the RFLPs of five control strains. When a total of 204 STEC O157 strains were analyzed by LA-PCR, one to three bands whose sizes ranged from 8.2 to 14 kb were obtained. Two STEC O157 strains, however, did not produce any bands. Subsequent restriction digest of the PCR products with BglI or EcoRV differentiated the RFLPs of 202 STEC O157 strains into 24 groups. The RFLP patterns of pulsed-field gel electrophoresis (PFGE) of representative strains of STEC O157 divided into 24 groups were well correlated with those of PCR-RFLP when STEC O157 strains were isolated in the same time period and in the close geographic area. To evaluate the PCR-RFLP assay developed here, ten strains, each isolated from four different outbreaks in different areas in Japan (Tochigi, Hyogo, Aichi, and Fukuoka prefecture), were examined to determine whether the strains in each group showed the same RFLP patterns in the PCR-RFLP assay. In accordance with the results of PFGE except for strains isolated in an area (Fukuoka), which did not produce any amplicon, ten strains in each group demonstrated the same RFLP pattern. Taken together, these data suggest that the PCR-RFLP based on region V is as useful as PFGE but perhaps more simple and rapid than PFGE for the molecular epidemiological analysis of STEC strains during sporadic and common source outbreaks.
Figures


Similar articles
-
Comparison of a PCR-restriction fragment length polymorphism (PCR-RFLP) assay to pulsed-field gel electrophoresis to determine the effect of repeated subculture and prolonged storage on RFLP patterns of Shiga toxin-producing Escherichia coli O157:H7.J Clin Microbiol. 2006 Nov;44(11):3963-8. doi: 10.1128/JCM.00717-06. Epub 2006 Sep 13. J Clin Microbiol. 2006. PMID: 16971640 Free PMC article.
-
Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina.Foodborne Pathog Dis. 2006 Spring;3(1):88-96. doi: 10.1089/fpd.2006.3.88. Foodborne Pathog Dis. 2006. PMID: 16602984
-
Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France.Appl Environ Microbiol. 2001 Jun;67(6):2460-8. doi: 10.1128/AEM.67.6.2460-2468.2001. Appl Environ Microbiol. 2001. PMID: 11375151 Free PMC article.
-
Physiologic and molecular markers for detection of shiga toxin-producing Escherichia coli serotype O26 strains.Foodborne Pathog Dis. 2006 Summer;3(2):163-77. doi: 10.1089/fpd.2006.3.163. Foodborne Pathog Dis. 2006. PMID: 16761942 Review.
-
[Coagulase gene polymorphism in Staphylococcus aureus--a new epidemiologic marker].Immun Infekt. 1995 Feb;23(1):9-14. Immun Infekt. 1995. PMID: 7698815 Review. German.
Cited by
-
Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting.Appl Environ Microbiol. 2008 Nov;74(21):6606-15. doi: 10.1128/AEM.00985-08. Epub 2008 Sep 12. Appl Environ Microbiol. 2008. PMID: 18791027 Free PMC article.
-
Comparison of a PCR-restriction fragment length polymorphism (PCR-RFLP) assay to pulsed-field gel electrophoresis to determine the effect of repeated subculture and prolonged storage on RFLP patterns of Shiga toxin-producing Escherichia coli O157:H7.J Clin Microbiol. 2006 Nov;44(11):3963-8. doi: 10.1128/JCM.00717-06. Epub 2006 Sep 13. J Clin Microbiol. 2006. PMID: 16971640 Free PMC article.
-
Development of a simple latex agglutination assay for detection of shiga toxin-producing Escherichia coli (STEC) by using polyclonal antibody against STEC.Clin Vaccine Immunol. 2007 May;14(5):600-4. doi: 10.1128/CVI.00342-06. Epub 2007 Mar 7. Clin Vaccine Immunol. 2007. PMID: 17344348 Free PMC article. Clinical Trial.
-
Rapid and Simple Universal Escherichia coli Genotyping Method Based on Multiple-Locus Variable-Number Tandem-Repeat Analysis Using Single-Tube Multiplex PCR and Standard Gel Electrophoresis.Appl Environ Microbiol. 2019 Mar 6;85(6):e02812-18. doi: 10.1128/AEM.02812-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30610078 Free PMC article.
-
Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains.J Clin Microbiol. 2009 Sep;47(9):2888-94. doi: 10.1128/JCM.00792-09. Epub 2009 Jul 29. J Clin Microbiol. 2009. PMID: 19641072 Free PMC article.
References
-
- Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. - PubMed
-
- Avery, S. M., E. Liebana, C. A. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. - PMC - PubMed
-
- Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

