Exploring the energy landscape of GFP by single-molecule mechanical experiments
- PMID: 15531635
- PMCID: PMC528946
- DOI: 10.1073/pnas.0404549101
Exploring the energy landscape of GFP by single-molecule mechanical experiments
Abstract
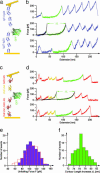
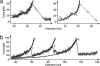
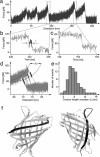
We use single-molecule force spectroscopy to drive single GFP molecules from the native state through their complex energy landscape into the completely unfolded state. Unlike many smaller proteins, mechanical GFP unfolding proceeds by means of two subsequent intermediate states. The transition from the native state to the first intermediate state occurs near thermal equilibrium at approximately 35 pN and is characterized by detachment of a seven-residue N-terminal alpha-helix from the beta barrel. We measure the equilibrium free energy cost associated with this transition as 22 k(B)T. Detachment of this small alpha-helix completely destabilizes GFP thermodynamically even though the beta-barrel is still intact and can bear load. Mechanical stability of the protein on the millisecond timescale, however, is determined by the activation barrier of unfolding the beta-barrel out of this thermodynamically unstable intermediate state. High bandwidth, time-resolved measurements of the cantilever relaxation phase upon unfolding of the beta-barrel revealed a second metastable mechanical intermediate with one complete beta-strand detached from the barrel. Quantitative analysis of force distributions and lifetimes lead to a detailed picture of the complex mechanical unfolding pathway through a rough energy landscape.
Figures



References
-
- Clausen-Schaumann, H., Seitz, M., Krautbauer, R. & Gaub, H. E. (2000) Curr. Opin. Chem. Biol. 4, 524-530. - PubMed
-
- Best, R. B. & Clarke, J. (2002) Chem. Commun. 183-192. - PubMed
-
- Fisher, T. E., Marszalek, P. E. & Fernandez, J. M. (2000) Nat. Struct. Biol. 7, 719-724. - PubMed
-
- Li, H., Linke, W. A., Oberhauser, A. F., Carrion-Vazquez, M., Kerkvliet, J. G., Lu, H., Marszalek, P. E. & Fernandez, J. M. (2002) Nature 418, 998-1002. - PubMed
-
- Schwaiger, I., Kardinal, A., Schleicher, M., Noegel, A. A. & Rief, M. (2004) Nat. Struct. Mol. Biol. 11, 81-85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

