Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae
- PMID: 15534003
- PMCID: PMC2172478
- DOI: 10.1083/jcb.200404173
Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae
Abstract
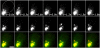
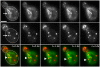
Using FM4-64 to label endosomes and Abp1p-GFP or Sac6p-GFP to label actin patches, we find that (1) endosomes colocalize with actin patches as they assemble at the bud cortex; (2) endosomes colocalize with actin patches as they undergo linear, retrograde movement from buds toward mother cells; and (3) actin patches interact with and disassemble at FM4-64-labeled internal compartments. We also show that retrograde flow of actin cables mediates retrograde actin patch movement. An Arp2/3 complex mutation decreases the frequency of cortical, nonlinear actin patch movements, but has no effect on the velocity of linear, retrograde actin patch movement. Rather, linear actin patch movement occurs at the same velocity and direction as the movement of actin cables. Moreover, actin patches require actin cables for retrograde movements and colocalize with actin cables as they undergo retrograde movement. Our studies support a mechanism whereby actin cables serve as "conveyor belts" for retrograde movement and delivery of actin patches/endosomes to FM4-64-labeled internal compartments.
Figures








References
-
- Ayscough, K.R. 2000. Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr. Biol. 10:1587–1590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

