Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment
- PMID: 15534202
- PMCID: PMC528977
- DOI: 10.1073/pnas.0407359101
Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment
Abstract
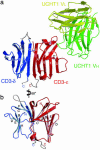
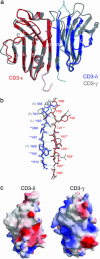
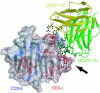
The alpha/beta T cell receptor complex transmits signals from MHC/peptide antigens through a set of constitutively associated signaling molecules, including CD3-epsilon/gamma and CD3-epsilon/delta. We report the crystal structure at 1.9-A resolution of a complex between a human CD3-epsilon/delta ectodomain heterodimer and a single-chain fragment of the UCHT1 antibody. CD3-epsilon/delta and CD3-epsilon/gamma share a conserved interface between the Ig-fold ectodomains, with parallel packing of the two G strands. CD3-delta has a more electronegative surface and a more compact Ig fold than CD3-gamma; thus, the two CD3 heterodimers have distinctly different molecular surfaces. The UCHT1 antibody binds near an acidic region of CD3-epsilon opposite the dimer interface, occluding this region from direct interaction with the TCR. This immunodominant epitope may be a uniquely accessible surface in the TCR/CD3 complex, because there is overlap between the binding site of the UCHT1 and OKT3 antibodies. Determination of the CD3-epsilon/delta structure completes the set of TCR/CD3 globular ectodomains and contributes information about exposed CD3 surfaces.
Figures


References
-
- Davis, M. M. & Bjorkman, P. J. (1988) Nature 334, 395-402. - PubMed
-
- Clevers, H., Alarcon, B., Wileman, T. & Terhorst, C. (1988) Annu. Rev. Immunol. 6, 629-662. - PubMed
-
- Letourneur, F. & Klausner, R. D. (1992) Science 255, 79-82. - PubMed
-
- Koning, F., Maloy, W. L. & Coligan, J. E. (1990) Eur. J. Immunol. 20, 299-305. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

