Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12
- PMID: 15534208
- PMCID: PMC526197
- DOI: 10.1073/pnas.0405821101
Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12
Abstract
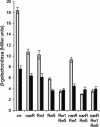
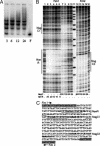
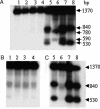
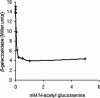
Bacterial-host attachment by means of bacterial adhesins is a key step in host colonization. Phase variation (reversible on-off switching) of the type 1 fimbrial adhesin of Escherichia coli involves a DNA inversion catalyzed by FimB (switching in either direction) or FimE (mainly on-to-off switching). fimB is separated from the divergent yjhATS operon by a large (1.4 kbp) intergenic region. Short ( approximately 28 bp) cis-active elements (regions 1 and 2) close to yjhA stimulate fimB expression and are required for sialic acid (Neu(5)Ac) sensitivity of its expression [El-Labany, S., Sohanpal, B. K., Lahooti, M., Akerman, R. & Blomfield, I. C. (2003) Mol. Microbiol. 49, 1109-1118]. Here, we show that whereas NanR, a sialic acid-response regulator, binds to region 1, NagC, a GlcNAc-6P-responsive protein, binds to region 2 instead. The NanR- and NagC-binding sites lie adjacent to deoxyadenosine methylase (Dam) methylation sites (5'-GATC) that are protected from modification, and the two regulators are shown to be required for methylation protection at regions 1 and 2, respectively. Mutations in nanR and nagC diminish fimB expression, and both fimB expression and FimB recombination are inhibited by GlcNAc (3- and >35-fold, respectively). Sialic acid catabolism generates GlcNAc-6-P, and whereas GlcNAc disrupts methylation protection by NagC alone, Neu(5)Ac inhibits the protection mediated by both NanR and NagC as expected. Type 1 fimbriae are proinflammatory, and host defenses enhance the release of both Neu(5)Ac and GlcNAc by a variety of mechanisms. Inhibition of type 1 fimbriation by these amino sugars may thus help balance the interaction between E. coli and its hosts.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

