Remote hot spots mediate protein substrate recognition for the Cdc25 phosphatase
- PMID: 15534213
- PMCID: PMC534539
- DOI: 10.1073/pnas.0407663101
Remote hot spots mediate protein substrate recognition for the Cdc25 phosphatase
Abstract
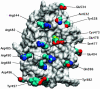
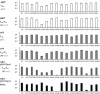
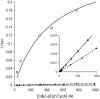
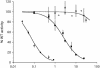
Cdc25B is a phosphatase that catalyzes the dephosphorylation and activation of the cyclin-dependent kinases, thus driving cell cycle progression. We have identified two residues, R488 and Y497, located >20 A from the active site, that mediate protein substrate recognition without affecting activity toward small-molecule substrates. Injection of Cdc25B wild-type but not the R488L or Y497A variants induces germinal vesicle breakdown and cyclin-dependent kinase activation in Xenopus oocytes. The conditional knockout of the cdc25 homolog (mih1) in Saccharomyces cerevisiae can be complemented by the wild type but not by the hot spot variants, indicating that protein substrate recognition by the Cdc25 phosphatases is an essential and evolutionarily conserved feature.
Figures


Similar articles
-
Redox control of cell cycle progression via Cdc25 phosphatase (Mih1p) in S. cerevisiae.Cell Cycle. 2006 Sep;5(18):2172-3. doi: 10.4161/cc.5.18.3252. Epub 2006 Sep 15. Cell Cycle. 2006. PMID: 16969116 No abstract available.
-
Crystal structure of Saccharomyces cerevisiae Ygr203w, a homolog of single-domain rhodanese and Cdc25 phosphatase catalytic domain.Proteins. 2009 Aug 1;76(2):520-4. doi: 10.1002/prot.22420. Proteins. 2009. PMID: 19382206 No abstract available.
-
Redundant Regulation of Cdk1 Tyrosine Dephosphorylation in Saccharomyces cerevisiae.Genetics. 2016 Mar;202(3):903-10. doi: 10.1534/genetics.115.182469. Epub 2015 Dec 29. Genetics. 2016. PMID: 26715668 Free PMC article.
-
Structure and function of the protein tyrosine phosphatases.Trends Biochem Sci. 1996 Nov;21(11):413-7. doi: 10.1016/s0968-0004(96)10059-1. Trends Biochem Sci. 1996. PMID: 8987394 Review.
-
Small molecule inhibitors of dual specificity protein phosphatases.Oncogene. 2000 Dec 27;19(56):6607-12. doi: 10.1038/sj.onc.1204084. Oncogene. 2000. PMID: 11426646 Review.
Cited by
-
CDC25B overexpression stabilises centrin 2 and promotes the formation of excess centriolar foci.PLoS One. 2013 Jul 1;8(7):e67822. doi: 10.1371/journal.pone.0067822. Print 2013. PLoS One. 2013. PMID: 23840880 Free PMC article.
-
Phosphatases and kinases regulating CDC25 activity in the cell cycle: clinical implications of CDC25 overexpression and potential treatment strategies.Mol Cell Biochem. 2016 May;416(1-2):33-46. doi: 10.1007/s11010-016-2693-2. Epub 2016 Apr 2. Mol Cell Biochem. 2016. PMID: 27038604 Free PMC article. Review.
-
Oncogenic Ras suppresses Cdk1 in a complex manner during the incubation of activated Xenopus egg extracts.Arch Biochem Biophys. 2013 Apr 15;532(2):61-72. doi: 10.1016/j.abb.2013.01.006. Epub 2013 Jan 29. Arch Biochem Biophys. 2013. PMID: 23376039 Free PMC article.
-
Phosphatases in Mitosis: Roles and Regulation.Biomolecules. 2019 Feb 7;9(2):55. doi: 10.3390/biom9020055. Biomolecules. 2019. PMID: 30736436 Free PMC article. Review.
-
The energetic network of hotspot residues between Cdc25B phosphatase and its protein substrate.J Mol Biol. 2006 Oct 6;362(5):1060-71. doi: 10.1016/j.jmb.2006.07.090. Epub 2006 Aug 15. J Mol Biol. 2006. PMID: 16950393 Free PMC article.
References
-
- Bogan, A. A. & Thorn, K. S. (1998) J. Mol. Biol. 280, 1–9. - PubMed
-
- Clackson, T. & Wells, J. A. (1995) Science 267, 383–386. - PubMed
-
- Schreiber, S. & Fersht, A. R. (1995) J. Mol. Biol. 248, 478–486. - PubMed
-
- Nilsson, I. & Hoffmann, I. (2000) Prog. Cell Cycle Res. 4, 107–114. - PubMed
-
- Kristjánsdóttir, K. & Rudolph, J. (2004) Chem. Biol. 11, 1043–1051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

