Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum
- PMID: 15535864
- PMCID: PMC545779
- DOI: 10.1186/gb-2004-5-11-r88
Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum
Abstract
Background: The apicomplexan parasite Cryptosporidium parvum is an emerging pathogen capable of causing illness in humans and other animals and death in immunocompromised individuals. No effective treatment is available and the genome sequence has recently been completed. This parasite differs from other apicomplexans in its lack of a plastid organelle, the apicoplast. Gene transfer, either intracellular from an endosymbiont/donor organelle or horizontal from another organism, can provide evidence of a previous endosymbiotic relationship and/or alter the genetic repertoire of the host organism. Given the importance of gene transfers in eukaryotic evolution and the potential implications for chemotherapy, it is important to identify the complement of transferred genes in Cryptosporidium.
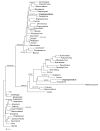
Results: We have identified 31 genes of likely plastid/endosymbiont (n = 7) or prokaryotic (n = 24) origin using a phylogenomic approach. The findings support the hypothesis that Cryptosporidium evolved from a plastid-containing lineage and subsequently lost its apicoplast during evolution. Expression analyses of candidate genes of algal and eubacterial origin show that these genes are expressed and developmentally regulated during the life cycle of C. parvum.
Conclusions: Cryptosporidium is the recipient of a large number of transferred genes, many of which are not shared by other apicomplexan parasites. Genes transferred from distant phylogenetic sources, such as eubacteria, may be potential targets for therapeutic drugs owing to their phylogenetic distance or the lack of homologs in the host. The successful integration and expression of the transferred genes in this genome has changed the genetic and metabolic repertoire of the parasite.
Figures





References
-
- CDC: bioterrorism agents/diseases http://www.bt.cdc.gov/agent/agentlist.asp
-
- Zhu G, Keithly JS, Philippe H. What is the phylogenetic position of Cryptosporidium? Int J Syst Evol Microbiol. 2000;50:1673–1681. - PubMed
-
- Carreno RA, Martin DS, Barta JR. Cryptosporidium is more closely related to the gregarines than to coccidia as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitol Res. 1999;85:899–904. doi: 10.1007/s004360050655. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

