Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana
- PMID: 15535868
- PMCID: PMC545783
- DOI: 10.1186/gb-2004-5-11-r92
Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana
Abstract
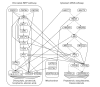
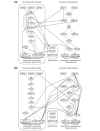
We present a novel graphical Gaussian modeling approach for reverse engineering of genetic regulatory networks with many genes and few observations. When applying our approach to infer a gene network for isoprenoid biosynthesis in Arabidopsis thaliana, we detect modules of closely connected genes and candidate genes for possible cross-talk between the isoprenoid pathways. Genes of downstream pathways also fit well into the network. We evaluate our approach in a simulation study and using the yeast galactose network.
Figures
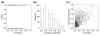
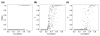

References
-
- Kurata H, El-Samad H, Yi TM, Khammash MJD. Feedback regulation of the heat shock response in E. coli. Proc 40th IEEE Conf Decision Control. 2001. pp. 837–842.
-
- Edwards D. Introduction to Graphical Modelling. 2. New York; Springer Verlag; 2000.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

