The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases
- PMID: 15535874
- PMCID: PMC545772
- DOI: 10.1186/gb-2004-5-11-248
The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases
Abstract
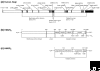
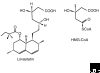
The enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase catalyzes the conversion of HMG-CoA to mevalonate, a four-electron oxidoreduction that is the rate-limiting step in the synthesis of cholesterol and other isoprenoids. The enzyme is found in eukaryotes and prokaryotes; and phylogenetic analysis has revealed two classes of HMG-CoA reductase, the Class I enzymes of eukaryotes and some archaea and the Class II enzymes of eubacteria and certain other archaea. Three-dimensional structures of the catalytic domain of HMG-CoA reductases from humans and from the bacterium Pseudomonas mevalonii, in conjunction with site-directed mutagenesis studies, have revealed details of the mechanism of catalysis. The reaction catalyzed by human HMG-CoA reductase is a target for anti-hypercholesterolemic drugs (statins), which are intended to lower cholesterol levels in serum. Eukaryotic forms of the enzyme are anchored to the endoplasmic reticulum, whereas the prokaryotic enzymes are soluble. Probably because of its critical role in cellular cholesterol homeostasis, mammalian HMG-CoA reductase is extensively regulated at the transcriptional, translational, and post-translational levels.
Figures

References
-
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. A study of the regulation of both mevalonate and mevalonate independent pathways for isoprenoid synthesis in plants. - DOI - PMC - PubMed
-
- Bochar DA, Stauffacher CV, Rodwell VW. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Genet Metab. 1999;66:122–127. doi: 10.1006/mgme.1998.2786. This article reported the classification of HMG-CoA reductases into Class I and Class II enzymes on the basis of sequence comparison. The authors utilized phylogenetic analysis to analyze a plethora of genomic sequences of various organisms. - DOI - PubMed
-
- Hedl M, Tabernero L, Stauffacher CV, Rodwell VW. Class II 3-hydroxy-3-methylglutaryl coenzyme A reductases. J Bacteriol. 2004;186:1927–1932. doi: 10.1128/JB.186.7.1927-1932.2004. A review article detailing current research and thought concerning Class II forms of the enzyme, including the HMGRs of many pathogenic bacteria. - DOI - PMC - PubMed
-
- Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. This article and [5] reported the crystal structure of the human HMG-CoA reductase catalytic domain, providing numerous insights into catalysis by a Class I HMG-CoA reductase. - DOI - PMC - PubMed
-
- Istvan ES, Deisenhofer J. The structure of the catalytic portion of human HMG-CoA reductase. Biochim Biophys Acta. 2000;1529:9–18. See [4] - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

