Molecular determinants of ligand selectivity in a vertebrate odorant receptor
- PMID: 15537883
- PMCID: PMC6730175
- DOI: 10.1523/JNEUROSCI.3117-04.2004
Molecular determinants of ligand selectivity in a vertebrate odorant receptor
Abstract
The identification of the chemical structure of an odorant by the vertebrate olfactory system is thought to occur through the combinatorial activity from multiple receptors, each tuned to recognize different chemical features. What are the molecular determinants underlying the selectivity of individual odorant receptors for their cognate ligands? To address this question, we performed molecular modeling and site-directed mutagenesis on the ligand-binding region of two orthologous amino acid odorant receptors belonging to the "C family" of G-protein-coupled receptors in goldfish and zebrafish. We identified the critical ligand-receptor interactions that afford ligand binding as well as selectivity for different amino acids. Moreover, predictions regarding binding pocket structure allowed us to alter, in a predictable manner, the receptor preferences for different ligands. These results reveal how this class of odorant receptor has evolved to accommodate ligands of varying chemical structure and further illuminate the molecular principles underlying ligand recognition and selectivity in this family of chemosensory receptors.
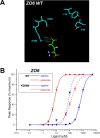
Figures




References
-
- Araneda RC, Kini AD, Firestein S (2000) The molecular receptive range of an odorant receptor. Nat Neurosci 3: 1248-1255. - PubMed
-
- Armstrong N, Sun Y, Chen GQ, Gouaux E (1998) Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395: 913-917. - PubMed
-
- Barth AL, Dugas JC, Ngai J (1997) Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron 19: 359-369. - PubMed
-
- Bertrand HO, Bessis AS, Pin JP, Acher FC (2002) Common and selective molecular determinants involved in metabotropic glutamate receptor agonist activity. J Med Chem 45: 3171-3183. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
