FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells
- PMID: 15539468
- PMCID: PMC535871
- DOI: 10.1105/tpc.104.027466
FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells
Abstract
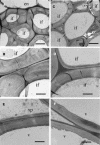
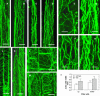
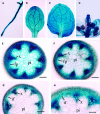
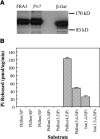
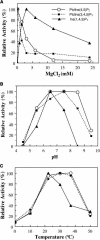
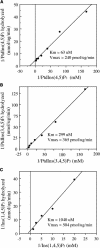
Type II inositol polyphosphate 5-phosphatases (5PTases) in yeast and animals have been known to regulate the level of phosphoinositides and thereby influence various cellular activities, such as vesicle trafficking and actin organization. In plants, little is known about the phosphatases involved in hydrolysis of phosphoinositides, and roles of type II 5PTases in plant cellular functions have not yet been characterized. In this study, we demonstrate that the FRAGILE FIBER3 (FRA3) gene of Arabidopsis thaliana, which encodes a type II 5PTase, plays an essential role in the secondary wall synthesis in fiber cells and xylem vessels. The fra3 mutations caused a dramatic reduction in secondary wall thickness and a concomitant decrease in stem strength. These phenotypes were associated with an alteration in actin organization in fiber cells. Consistent with the defective fiber and vessel phenotypes, the FRA3 gene was found to be highly expressed in fiber cells and vascular tissues in stems. The FRA3 protein is composed of two domains, an N-terminal localized WD-repeat domain and a C-terminal localized 5PTase catalytic domain. In vitro activity assay demonstrated that recombinant FRA3 exhibited phosphatase activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3, with the highest substrate affinity toward PtdIns(4,5)P2. The fra3 missense mutation, which caused an amino acid substitution in the conserved motif II of the 5PTase catalytic domain, completely abolished the FRA3 phosphatase activity. Moreover, the endogenous levels of PtdIns(4,5)2 and Ins(1,4,5)P3 were found to be elevated in fra3 stems. Together, our findings suggest that the FRA3 type II 5PTase is involved in phosphoinositide metabolism and influences secondary wall synthesis and actin organization.
Figures






References
-
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus, and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
-
- Despres, B., Bouissonnié, F., Wu, H.-J., Gomord, V., Guilleminot, J., Grellet, F., Berger, F., Delseny, M., and Devic, M. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

