Insights into saquinavir resistance in the G48V HIV-1 protease: quantum calculations and molecular dynamic simulations
- PMID: 15542562
- PMCID: PMC1305161
- DOI: 10.1529/biophysj.104.046110
Insights into saquinavir resistance in the G48V HIV-1 protease: quantum calculations and molecular dynamic simulations
Abstract
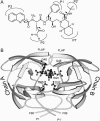
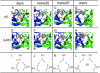
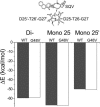
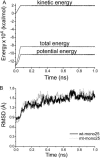
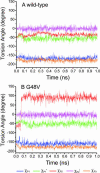
The spread of acquired immune deficiency syndrome has increasingly become a great concern owing largely to the failure of chemotherapies. The G48V is considered the key signature residue mutation of HIV-1 protease developing with saquinavir therapy. Molecular dynamics simulations of the wild-type and the G48V HIV-1 protease complexed with saquinavir were carried out to explore structure and interactions of the drug resistance. The molecular dynamics results combined with the quantum-based and molecular mechanics Poisson-Boltzmann surface area calculations indicated a monoprotonation took place on D25, one of the triad active site residues. The inhibitor binding of the triad residues and its interaction energy in the mutant were similar to those in the wild-type. The overall structure of both complexes is almost identical. However, the steric conflict of the substituted valine results in the conformational change of the P2 subsite and the disruption of hydrogen bonding between the -NH of the P2 subsite and the backbone -CO of the mutated residue. The magnitude of interaction energy changes was comparable to the experimental K(i) data. The designing for a new drug should consider a reduction of steric repulsion on P2 to enhance the activity toward this mutant strain.
Figures
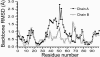

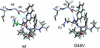

Similar articles
-
Structural and dynamical properties of different protonated states of mutant HIV-1 protease complexed with the saquinavir inhibitor studied by molecular dynamics simulations.J Mol Graph Model. 2006 Nov;25(3):324-32. doi: 10.1016/j.jmgm.2006.01.004. Epub 2006 Feb 28. J Mol Graph Model. 2006. PMID: 16504560
-
Insight into analysis of interactions of saquinavir with HIV-1 protease in comparison between the wild-type and G48V and G48V/L90M mutants based on QM and QM/MM calculations.J Mol Graph Model. 2007 Nov;26(4):720-7. doi: 10.1016/j.jmgm.2007.04.009. Epub 2007 May 3. J Mol Graph Model. 2007. PMID: 17543558
-
Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance.Protein Sci. 2000 Oct;9(10):1898-904. doi: 10.1110/ps.9.10.1898. Protein Sci. 2000. PMID: 11106162 Free PMC article.
-
Probing structure-function relationships in human immunodeficiency virus type 1 protease via molecular dynamics simulation.Methods Enzymol. 1994;241:178-95. doi: 10.1016/0076-6879(94)41065-8. Methods Enzymol. 1994. PMID: 7854178 Review.
-
Rational approaches to resistance: using saquinavir.AIDS. 1996 Nov;10 Suppl 1:S15-9. AIDS. 1996. PMID: 8970671 Review.
Cited by
-
Perspectives towards antiviral drug discovery against Ebola virus.J Med Virol. 2019 Dec;91(12):2029-2048. doi: 10.1002/jmv.25357. Epub 2019 Sep 30. J Med Virol. 2019. PMID: 30431654 Free PMC article. Review.
-
Mechanism of drug resistance in HIV-1 protease subtype C in the presence of Atazanavir.Curr Res Struct Biol. 2024 Feb 20;7:100132. doi: 10.1016/j.crstbi.2024.100132. eCollection 2024. Curr Res Struct Biol. 2024. PMID: 38435053 Free PMC article.
-
Identification of potential inhibitors based on compound proposal contest: Tyrosine-protein kinase Yes as a target.Sci Rep. 2015 Nov 26;5:17209. doi: 10.1038/srep17209. Sci Rep. 2015. PMID: 26607293 Free PMC article.
-
FMO-guided design of darunavir analogs as HIV-1 protease inhibitors.Sci Rep. 2024 Feb 13;14(1):3639. doi: 10.1038/s41598-024-53940-1. Sci Rep. 2024. PMID: 38351065 Free PMC article.
-
Evaluating the potency of HIV-1 protease drugs to combat resistance.Proteins. 2008 May 15;71(3):1163-74. doi: 10.1002/prot.21808. Proteins. 2008. PMID: 18004760 Free PMC article.
References
-
- Antosiewicz, J., J. A. Mccammon, and M. K. Gilson. 1994. Prediction of pH-dependent properties of proteins. J. Mol. Biol. 238:415–436. - PubMed
-
- Baldwin, E. T., T. N. Bhat, S. Gulnik, B. Liu, I. A. Topol, Y. Kiso, T. Mimoto, H. Mitsuya, and J. W. Erickson. 1995. Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine. Structure. 3:581–590. - PubMed
-
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690.
-
- Boucher, C. 1996. Rational approaches to resistance: using saquinavir. AIDS. 10 (Suppl. 1) :S15–S19. - PubMed
-
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem. 4:187–217.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

