Multiple genotypes of influenza B virus circulated between 1979 and 2003
- PMID: 15542634
- PMCID: PMC525012
- DOI: 10.1128/JVI.78.23.12817-12828.2004
Multiple genotypes of influenza B virus circulated between 1979 and 2003
Erratum in
- J Virol. 2005 May;79(9):5886
Abstract
The segmented genome of influenza B virus allows exchange of gene segments between cocirculating strains. Through this process of reassortment, diversity is generated by the mixing of genes between viruses that differ in one or more gene segments. Phylogenetic and evolutionary analyses of all 11 genes of 31 influenza B viruses isolated from 1979 to 2003 were used to study the evolution of whole genomes. All 11 genes diverged into two new lineages prior to 1987. All genes except the NS1 gene were undergoing linear evolution, although the rate of evolution and the degree to which nucleotide changes translated into amino acid changes varied between lineages and by gene. Frequent reassortment generated 14 different genotypes distinct from the gene constellation of viruses circulating prior to 1979. Multiple genotypes cocirculated in some locations, and a sequence of reassortment events over time could not be established. The surprising diversity of the viruses, unrestricted mixing of lineages, and lack of evidence for coevolution of gene segments do not support the hypothesis that the reassortment process is driven by selection for functional differences.
Figures
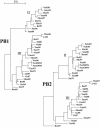
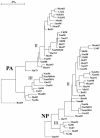
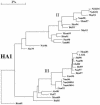
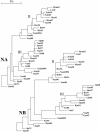
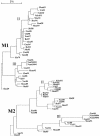
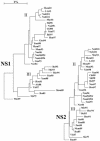

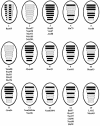
References
-
- Air, G. M., W. G. Laver, M. Luo, S. J. Stray, G. Legrone, and R. G. Webster. 1990. Antigenic, sequence, and crystal variation in influenza B neuraminidase. Virology 177:578-587. - PubMed
-
- Akoto-Amanfu, E., N. Sivasubramanian, and D. P. Nayak. 1987. Primary structure of the polymerase acidic (PA) gene of an influenza B virus (B/Sing/222/79). Virology 159:147-153. - PubMed
-
- Barr, I. G., N. Komadina, A. Hurt, R. Shaw, C. Durrant, P. Iannello, C. Tomasov, H. Sjogren, and A. W. Hampson. 2003. Reassortants in recent human influenza A and B isolates from South East Asia and Oceania. Virus Res. 98:35-44. - PubMed
-
- Burmeister, W. P., F. Baudin, S. Cusack, and R. W. Ruigrok. 1993. Comparison of structure and sequence of influenza B/Yamagata and B/Beijing neuraminidases shows a conserved “head” but much greater variability in the “stalk” and NB protein. Virology 192:683-686. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

