Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis
- PMID: 15542639
- PMCID: PMC524998
- DOI: 10.1128/JVI.78.23.12868-12876.2004
Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis
Abstract
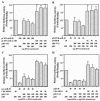
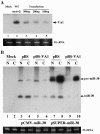
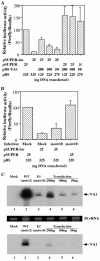
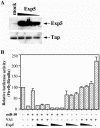
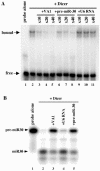
Although inhibition of RNA interference (RNAi) by plant virus proteins has been shown to enhance viral replication and pathogenesis in plants, no viral gene product has as yet been shown to inhibit RNAi in vertebrate cells. Here, we present evidence demonstrating that the highly structured approximately 160-nucleotide adenoviral VA1 noncoding RNA can inhibit RNAi at physiological levels of expression. VA1, which is expressed at very high levels in adenovirus-infected cells, potently inhibited RNAi induced by short hairpin RNAs (shRNAs) or human microRNA precursors but did not affect RNAi induced by artificial short interfering RNA duplexes. Inhibition appeared to be due both to inhibition of nuclear export of shRNA or premicro-RNA precursors, competition for the Exportin 5 nuclear export factor, and inhibition of Dicer function by direct binding of Dicer. Together, these data argue that adenovirus infection can result in inhibition of RNAi and identify VA1 RNA as the first viral gene product able to inhibit RNAi in human cells.
Figures

References
-
- Arts, G.-J., E. Langemeijer, R. Tissingh, L. Ma, H. Pavliska, K. Dokic, R. Dooijes, E. Mešić, R. Clasen, F. Michiels, J. van der Schueren, M. Lambrecht, S. Herman, R. Brys, K. Thys, M. Hoffmann, P. Tomme, and H. van Es. 2003. Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res. 13:2325-2332. - PMC - PubMed
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. - PubMed
-
- Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:295-296. - PubMed
-
- Bridge, A. J., S. Pebernard, A. Ducraux, A.-L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. - PubMed
-
- Brummelkamp, T., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

