Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies
- PMID: 15542675
- PMCID: PMC524984
- DOI: 10.1128/JVI.78.23.13232-13252.2004
Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies
Abstract
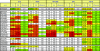
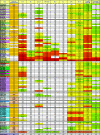
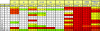
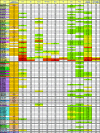
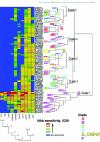
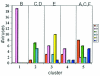
Broadly neutralizing monoclonal antibodies (MAbs) are potentially important tools in human immunodeficiency virus type 1 (HIV-1) vaccine design. A few rare MAbs have been intensively studied, but we still have a limited appreciation of their neutralization breadth. Using a pseudovirus assay, we evaluated MAbs from clade B-infected donors and a clade B HIV(+) plasma against 93 viruses from diverse backgrounds. Anti-gp120 MAbs exhibited greater activity against clade B than non-B viruses, whereas anti-gp41 MAbs exhibited broad interclade activity. Unexpectedly, MAb 4E10 (directed against the C terminus of the gp41 ectodomain) neutralized all 90 viruses with moderate potency. MAb 2F5 (directed against an epitope adjacent to that of 4E10) neutralized 67% of isolates, but none from clade C. Anti-gp120 MAb b12 (directed against an epitope overlapping the CD4 binding site) neutralized 50% of viruses, including some from almost every clade. 2G12 (directed against a high-mannose epitope on gp120) neutralized 41% of the viruses, but none from clades C or E. MAbs to the gp120 V3 loop, including 447-52D, neutralized a subset of clade B viruses (up to 45%) but infrequently neutralized other clades (</=7%). MAbs b6 (directed against the CD4 binding site) and X5 (directed against a CD4-induced epitope of gp120) neutralized only sensitive primary clade B viruses. The HIV(+) plasma neutralized 70% of the viruses, including some from all major clades. Further analysis revealed five neutralizing immunotypes that were somewhat associated with clades. As well as the significance for vaccine design, our data have implications for passive-immunization studies in countries where clade C viruses are common, given that only MAbs b12 and 4E10 were effective against viruses from this clade.
Figures




References
-
- Allaway, G. P., A. M. Ryder, G. A. Beaudry, and P. J. Maddon. 1993. Synergistic inhibition of HIV-1 envelope-mediated cell fusion by CD4-based molecules in combination with antibodies to gp120 or gp41. AIDS Res. Hum. Retrovir. 9:581-587. - PubMed
-
- Anonymous.1994. HIV type 1 variation in World Health Organization-sponsored vaccine evaluation sites: genetic screening, sequence analysis, and preliminary biological characterization of selected viral strains. AIDS Res. Hum. Retrovir. 10:1327-1343. - PubMed
-
- Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

