Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors
- PMID: 15542826
- PMCID: PMC529025
- DOI: 10.1128/MCB.24.23.10151-10160.2004
Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors
Abstract
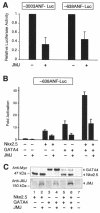
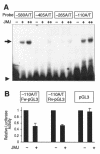
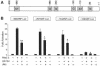
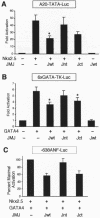
Mice with a homozygous knockout of the jumonji (jmj) gene showed abnormal heart development and defective regulation of cardiac-specific genes, including the atrial natriuretic factor (ANF). ANF is one of the earliest markers of cardiac differentiation and a hallmark for cardiac hypertrophy. Here, we show that JMJ represses ANF gene expression by inhibiting transcriptional activities of Nkx2.5 and GATA4. JMJ represses the Nkx2.5- or GATA4-dependent activation of the reporter genes containing the ANF promoter-enhancer or containing the Nkx2.5 or GATA4-binding consensus sequence. JMJ physically associates with Nkx2.5 and GATA4 in vitro and in vivo as determined by glutathione S-transferase pull-down and immunoprecipitation assays. Using mutational analyses, we mapped the protein-protein interaction domains in JMJ, Nkx2.5, and GATA4. We identified two DNA-binding sites of JMJ in the ANF enhancer by gel mobility shift assays. However, these JMJ-binding sites do not seem to mediate ANF repression by JMJ. Mutational analysis of JMJ indicates that the protein-protein interaction domain of JMJ mediates the repression of ANF gene expression. Therefore, JMJ may play important roles in the down-regulation of ANF gene expression and in heart development.
Figures




References
-
- Balciunas, D., and H. Ronne. 2000. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25:274-276. - PubMed
-
- Bao, Z. Z., B. G. Bruneau, J. G. Seidman, C. E. Seidman, and C. L. Cepko. 1999. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283:1161-1164. - PubMed
-
- Bruneau, B. G., G. Nemer, J. P. Schmitt, F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709-721. - PubMed
-
- Chen, F., H. Kook, R. Milewski, A. D. Gitler, M. M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, G. Runke, J. P. Mackay, J. Novotny, R. J. Schwartz, R. P. Harvey, M. C. Mullins, and J. A. Epstein. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713-723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
