Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2
- PMID: 15542827
- PMCID: PMC529034
- DOI: 10.1128/MCB.24.23.10161-10168.2004
Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2
Abstract
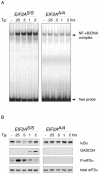
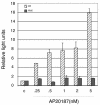
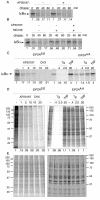
Numerous stressful conditions activate kinases that phosphorylate the alpha subunit of translation initiation factor 2 (eIF2alpha), thus attenuating mRNA translation and activating a gene expression program known as the integrated stress response. It has been noted that conditions associated with eIF2alpha phosphorylation, notably accumulation of unfolded proteins in the endoplasmic reticulum (ER), or ER stress, are also associated with activation of nuclear factor kappa B (NF-kappaB) and that eIF2alpha phosphorylation is required for NF-kappaB activation by ER stress. We have used a pharmacologically activable version of pancreatic ER kinase (PERK, an ER stress-responsive eIF2alpha kinase) to uncouple eIF2alpha phosphorylation from stress and found that phosphorylation of eIF2alpha is both necessary and sufficient to activate both NF-kappaB DNA binding and an NF-kappaB reporter gene. eIF2alpha phosphorylation-dependent NF-kappaB activation correlated with decreased levels of the inhibitor IkappaBalpha protein. Unlike canonical signaling pathways that promote IkappaBalpha phosphorylation and degradation, eIF2alpha phosphorylation did not increase phosphorylated IkappaBalpha levels or affect the stability of the protein. Pulse-chase labeling experiments indicate instead that repression of IkappaBalpha translation plays an important role in NF-kappaB activation in cells experiencing high levels of eIF2alpha phosphorylation. These studies suggest a direct role for eIF2alpha phosphorylation-dependent translational control in activating NF-kappaB during ER stress.
Figures




Similar articles
-
GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation.Biochem J. 2005 Jan 15;385(Pt 2):371-80. doi: 10.1042/BJ20041164. Biochem J. 2005. PMID: 15355306 Free PMC article.
-
Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses.Mol Cell Biol. 2003 Aug;23(16):5651-63. doi: 10.1128/MCB.23.16.5651-5663.2003. Mol Cell Biol. 2003. PMID: 12897138 Free PMC article.
-
Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α.J Biol Chem. 2014 May 2;289(18):12593-611. doi: 10.1074/jbc.M113.543215. Epub 2014 Mar 19. J Biol Chem. 2014. PMID: 24648524 Free PMC article.
-
Coping with stress: eIF2 kinases and translational control.Biochem Soc Trans. 2006 Feb;34(Pt 1):7-11. doi: 10.1042/BST20060007. Biochem Soc Trans. 2006. PMID: 16246168 Review.
-
Bidirectional regulation of NF-κB by reactive oxygen species: a role of unfolded protein response.Free Radic Biol Med. 2013 Dec;65:162-174. doi: 10.1016/j.freeradbiomed.2013.06.020. Epub 2013 Jun 19. Free Radic Biol Med. 2013. PMID: 23792277 Review.
Cited by
-
Oxidative stress, unfolded protein response, and apoptosis in developmental toxicity.Int Rev Cell Mol Biol. 2015;317:1-66. doi: 10.1016/bs.ircmb.2015.02.002. Epub 2015 Mar 11. Int Rev Cell Mol Biol. 2015. PMID: 26008783 Free PMC article. Review.
-
High-fat diet aggravates colitis via mesenteric adipose tissue derived exosome metastasis-associated lung adenocarcinoma transcript 1.World J Gastroenterol. 2022 Aug 7;28(29):3838-3853. doi: 10.3748/wjg.v28.i29.3838. World J Gastroenterol. 2022. PMID: 36157545 Free PMC article.
-
Platelet factor 4 (CXCL4/PF4) upregulates matrix metalloproteinase-2 (MMP-2) in gingival fibroblasts.Sci Rep. 2022 Nov 3;12(1):18636. doi: 10.1038/s41598-022-19850-w. Sci Rep. 2022. PMID: 36329090 Free PMC article.
-
Cross-talk between the unfolded protein response and nuclear factor-κB signalling pathways regulates cytokine-mediated beta cell death in MIN6 cells and isolated mouse islets.Diabetologia. 2012 Nov;55(11):2999-3009. doi: 10.1007/s00125-012-2657-3. Epub 2012 Jul 28. Diabetologia. 2012. PMID: 22893028
-
Functional analysis of the short isoform of orf virus protein OV20.0.J Virol. 2015 May;89(9):4966-79. doi: 10.1128/JVI.03714-14. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694596 Free PMC article.
References
-
- Baeuerle, P. A., and D. Baltimore. 1988. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science 242:540-546. - PubMed
-
- Bertolotti, A., Y. Zhang, L. Hendershot, H. Harding, and D. Ron. 2000. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat. Cell Biol. 2:326-332. - PubMed
-
- Chen, J. 2000. Heme-regulated eIF2α kinase, p. 529-546. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. CSHL Press, Cold Spring Harbor, N.Y.
-
- Cheshire, J. L., B. R. Williams, and A. S. Baldwin, Jr. 1999. Involvement of double-stranded RNA-activated protein kinase in the synergistic activation of nuclear factor-kappaB by tumor necrosis factor-alpha and gamma-interferon in preneuronal cells. J. Biol. Chem. 274:4801-4806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
