Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity
- PMID: 15542830
- PMCID: PMC529035
- DOI: 10.1128/MCB.24.23.10193-10207.2004
Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity
Erratum in
- Mol Cell Biol. 2005 Feb;25(3):1213
Abstract
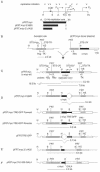
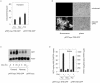
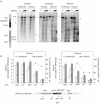
The observation that transcriptionally active genes generally replicate early in S phase and observations of the interaction between transcription factors and replication proteins support the thesis that promoter elements may have a role in DNA replication. To test the relationship between transcription and replication we constructed HeLa cell lines in which inducible green fluorescent protein (GFP)-encoding genes replaced the proximal approximately 820-bp promoter region of the c-myc gene. Without the presence of an inducer, basal expression occurred from the GFP gene in either orientation and origin activity was restored to the mutant c-myc replicator. In contrast, replication initiation was repressed upon induction of transcription. When basal or induced transcription complexes were slowed by the presence of alpha-amanitin, origin activity depended on the orientation of the transcription unit. To test mechanistically whether basal transcription or transcription factor binding was sufficient for replication rescue by the uninduced GFP genes, a GAL4p binding cassette was used to replace all regulatory sequences within approximately 1,400 bp 5' to the c-myc gene. In these cells, expression of a CREB-GAL4 fusion protein restored replication origin activity. These results suggest that transcription factor binding can enhance replication origin activity and that high levels of expression or the persistence of transcription complexes can repress it.
Figures




Similar articles
-
c-Myc creates an activation loop by transcriptionally repressing its own functional inhibitor, hMad4, in young fibroblasts, a loop lost in replicatively senescent fibroblasts.J Cell Biochem. 2005 Dec 1;96(5):1071-85. doi: 10.1002/jcb.20503. J Cell Biochem. 2005. PMID: 16167342
-
Basal components of the transcription apparatus (RNA polymerase II, TATA-binding protein) contain activation domains: is the repetitive C-terminal domain (CTD) of RNA polymerase II a "portable enhancer domain"?Mol Reprod Dev. 1994 Oct;39(2):215-25. doi: 10.1002/mrd.1080390215. Mol Reprod Dev. 1994. PMID: 7826625
-
TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression.Mol Cell Biol. 2005 Jan;25(1):147-61. doi: 10.1128/MCB.25.1.147-161.2005. Mol Cell Biol. 2005. PMID: 15601838 Free PMC article.
-
Making myc.Curr Top Microbiol Immunol. 2006;302:1-32. doi: 10.1007/3-540-32952-8_1. Curr Top Microbiol Immunol. 2006. PMID: 16620023 Review.
-
Gene expression regulation and cancer.Clin Transl Oncol. 2006 Nov;8(11):780-7. doi: 10.1007/s12094-006-0132-7. Clin Transl Oncol. 2006. PMID: 17134965 Review.
Cited by
-
Discrete functional elements required for initiation activity of the Chinese hamster dihydrofolate reductase origin beta at ectopic chromosomal sites.Exp Cell Res. 2007 Jan 1;313(1):109-20. doi: 10.1016/j.yexcr.2006.09.020. Epub 2006 Sep 28. Exp Cell Res. 2007. PMID: 17078947 Free PMC article.
-
Differential binding of replication proteins across the human c-myc replicator.Mol Cell Biol. 2006 Jul;26(14):5270-83. doi: 10.1128/MCB.02137-05. Mol Cell Biol. 2006. PMID: 16809765 Free PMC article.
-
Unstable spinocerebellar ataxia type 10 (ATTCT*(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells.Mol Cell Biol. 2007 Nov;27(22):7828-38. doi: 10.1128/MCB.01276-07. Epub 2007 Sep 10. Mol Cell Biol. 2007. PMID: 17846122 Free PMC article.
-
Human gene organization driven by the coordination of replication and transcription.Genome Res. 2007 Sep;17(9):1278-85. doi: 10.1101/gr.6533407. Epub 2007 Aug 3. Genome Res. 2007. PMID: 17675363 Free PMC article.
-
Decreased replication origin activity in temporal transition regions.J Cell Biol. 2009 Nov 30;187(5):623-35. doi: 10.1083/jcb.200905144. J Cell Biol. 2009. PMID: 19951913 Free PMC article.
References
-
- Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. - PubMed
-
- Archard, L. C., K. Johnson, and A. D. Malcolm. 1985. Specific transcription of orthopox virus DNA by HeLa cell RNA polymerase II. FEBS Lett. 192:53-56. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. Wiley, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
