Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein
- PMID: 15542854
- PMCID: PMC529055
- DOI: 10.1128/MCB.24.23.10470-10478.2004
Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein
Abstract
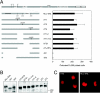
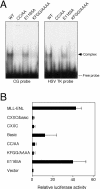
The MLL gene is a frequent target for leukemia-associated chromosomal translocations that generate dominant-acting chimeric oncoproteins. These invariably contain the amino-terminal 1,400 residues of MLL fused with one of a variety of over 30 distinct nuclear or cytoplasmic partner proteins. Despite the consistent inclusion of the MLL amino-terminal region in leukemia oncoproteins, little is known regarding its molecular contributions to MLL-dependent oncogenesis. Using high-resolution mutagenesis, we identified three MLL domains that are essential for in vitro myeloid transformation via mechanisms that do not compromise subnuclear localization. These include the CXXC/Basic domain and two novel domains of unknown function. Point mutations in the CXXC domain that eliminate myeloid transformation by an MLL fusion protein also abolished recognition and binding of nonmethylated CpG DNA sites in vitro and transactivation in vivo. Our results define a critical role for the CXXC DNA binding domain in MLL-associated oncogenesis, most likely via epigenetic recognition of CpG DNA sites within the regulatory elements of target genes.
Figures


References
-
- Armstrong, S. A., J. E. Staunton, L. B. Silverman, R. Pieters, M. L. den Boer, M. D. Minden, S. E. Sallan, E. S. Lander, T. R. Golub, and S. J. Korsmeyer. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30:41-47. - PubMed
-
- Ayton, P., S. F. Sneddon, D. B. Palmer, I. R. Rosewell, M. J. Owen, B. Young, R. Presley, and V. Subramanian. 2001. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis 30:201-212. - PubMed
-
- Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. - PubMed
-
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
