Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations
- PMID: 15543491
- PMCID: PMC1196432
- DOI: 10.1086/426952
Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations
Abstract
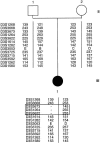
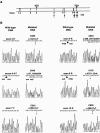
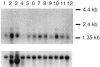
Cerebral cavernous malformations (CCMs) are hamartomatous vascular malformations characterized by abnormally enlarged capillary cavities without intervening brain parenchyma. They cause seizures and cerebral hemorrhages, which can result in focal neurological deficits. Three CCM loci have been mapped, and loss-of-function mutations were identified in the KRIT1 (CCM1) and MGC4607 (CCM2) genes. We report herein the identification of PDCD10 (programmed cell death 10) as the CCM3 gene. The CCM3 locus has been previously mapped to 3q26-27 within a 22-cM interval that is bracketed by D3S1763 and D3S1262. We hypothesized that genomic deletions might occur at the CCM3 locus, as reported previously to occur at the CCM2 locus. Through high-density microsatellite genotyping of 20 families, we identified, in one family, null alleles that resulted from a deletion within a 4-Mb interval flanked by markers D3S3668 and D3S1614. This de novo deletion encompassed D3S1763, which strongly suggests that the CCM3 gene lies within a 970-kb region bracketed by D3S1763 and D3S1614. Six additional distinct deleterious mutations within PDCD10, one of the five known genes mapped within this interval, were identified in seven families. Three of these mutations were nonsense mutations, and two led to an aberrant splicing of exon 9, with a frameshift and a longer open reading frame within exon 10. The last of the six mutations led to an aberrant splicing of exon 5, without frameshift. Three of these mutations occurred de novo. All of them cosegregated with the disease in the families and were not observed in 200 control chromosomes. PDCD10, also called "TFAR15," had been initially identified through a screening for genes differentially expressed during the induction of apoptosis in the TF-1 premyeloid cell line. It is highly conserved in both vertebrates and invertebrates. Its implication in cerebral cavernous malformations strongly suggests that it is a new player in vascular morphogenesis and/or remodeling.
Figures


References
Electronic-Database Information
-
- ExPASy Proteomics Server, http://us.expasy.org/ (for structural feature prediction)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequence information for 3q genomic contigs and the PDCD10 alternative transcripts [accession numbers NM_007217, NM_145859, and NM_145860])
-
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCM) - PubMed
-
- UniGene, http://www.ncbi.nlm.nih.gov/UniGene/ (for Mus musculus Pdcd10 [accession number Mm.316473])
References
-
- Craig HD, Gunel M, Cepeda O, Johnson EW, Ptacek L, Steinberg GK, Ogilvy CS, Berg MJ, Crawford SC, Scott RM, Sabroe R, Kennedy CT, Mettler G, Beis MJ, Fryer A, Awad IA, Lifton RP (1998) Multilocus linkage identifies two new loci for a Mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum Mol Genet 7:1851–185810.1093/hmg/7.12.1851 - DOI - PubMed
-
- Dubovsky J, Zabramski JM, Kurth J, Spetzler RF, Rich SS, Orr HT, Weber JL (1995) A gene responsible for cavernous malformations of the brain maps to chromosome 7q. Hum Mol Genet 4:453–458 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

