Coelacanth genome sequence reveals the evolutionary history of vertebrate genes
- PMID: 15545497
- PMCID: PMC534663
- DOI: 10.1101/gr.2972804
Coelacanth genome sequence reveals the evolutionary history of vertebrate genes
Abstract
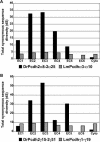
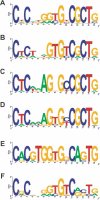
The coelacanth is one of the nearest living relatives of tetrapods. However, a teleost species such as zebrafish or Fugu is typically used as the outgroup in current tetrapod comparative sequence analyses. Such studies are complicated by the fact that teleost genomes have undergone a whole-genome duplication event, as well as individual gene-duplication events. Here, we demonstrate the value of coelacanth genome sequence by complete sequencing and analysis of the protocadherin gene cluster of the Indonesian coelacanth, Latimeria menadoensis. We found that coelacanth has 49 protocadherin cluster genes organized in the same three ordered subclusters, alpha, beta, and gamma, as the 54 protocadherin cluster genes in human. In contrast, whole-genome and tandem duplications have generated two zebrafish protocadherin clusters comprised of at least 97 genes. Additionally, zebrafish protocadherins are far more prone to homogenizing gene conversion events than coelacanth protocadherins, suggesting that recombination- and duplication-driven plasticity may be a feature of teleost genomes. Our results indicate that coelacanth provides the ideal outgroup sequence against which tetrapod genomes can be measured. We therefore present L. menadoensis as a candidate for whole-genome sequencing.
Figures
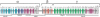





References
-
- Amores, A., Force, A., Yan, Y.L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. - PubMed
-
- Bailey, T. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the second international conference on intelligent systems for molecular biology, pp. 28-36, AAAI Press, Menlo Park, CA. - PubMed
Web site references
-
- http://lagan.stanford.edu/; LAGAN.
-
- http://hmmer.wustl.edu/; HMMER.
-
- http://www.cbs.dtu.dk/services/RevTrans/; RevTrans.
-
- http://weblogo.berkeley.edu/logo.cgi; WebLogo.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
