Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques
- PMID: 15545610
- PMCID: PMC534505
- DOI: 10.1073/pnas.0402766101
Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques
Abstract
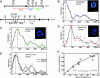
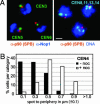
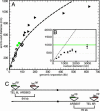
Little is known about how chromatin folds in its native state. Using optimized in situ hybridization and live imaging techniques have determined compaction ratios and fiber flexibility for interphase chromatin in budding yeast. Unlike previous studies, ours examines nonrepetitive chromatin at intervals short enough to be meaningful for yeast chromosomes and functional domains in higher eukaryotes. We reconcile high-resolution fluorescence in situ hybridization data from intervals of 14-100 kb along single chromatids with measurements of whole chromosome arms (122-623 kb in length), monitored in intact cells through the targeted binding of bacterial repressors fused to GFP derivatives. The results are interpreted with a flexible polymer model and suggest that interphase chromatin exists in a compact higher-order conformation with a persistence length of 170-220 nm and a mass density of approximately 110-150 bp/nm. These values are equivalent to 7-10 nucleosomes per 11-nm turn within a 30-nm-like fiber structure. Comparison of long and short chromatid arm measurements demonstrates that chromatin fiber extension is also influenced by nuclear geometry. The observation of this surprisingly compact chromatin structure for transcriptionally competent chromatin in living yeast cells suggests that the passage of RNA polymerase II requires a very transient unfolding of higher-order chromatin structure.
Figures

References
-
- Woodcock, C. L. & Dimitrov, S. (2001) Curr. Opin. Genet. Dev. 11, 130–135. - PubMed
-
- Hansen, J. C. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 361–392. - PubMed
-
- Gasser, S. M. (2002) Science 296, 1412–1416. - PubMed
-
- Marshall, W. F. (2002) Curr. Biol. 12, R185–R192. - PubMed
-
- van den Engh, G., Sachs, R. & Trask, B. J. (1992) Science 257, 1410–1412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

