Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]
- PMID: 15545639
- PMCID: PMC1449557
- DOI: 10.1534/genetics.104.037168
Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]
Abstract
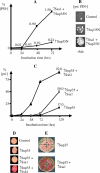
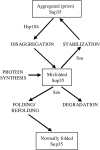
[PSI(+)] is a prion isoform of the yeast release factor Sup35. In some assays, the cytosolic chaperones Ssa1 and Ssb1/2 of the Hsp70 family were previously shown to exhibit "pro-[PSI(+)]" and "anti-[PSI(+)]" effects, respectively. Here, it is demonstrated for the first time that excess Ssa1 increases de novo formation of [PSI(+)] and that pro-[PSI(+)] effects of Ssa1 are shared by all other Ssa proteins. Experiments with chimeric constructs show that the peptide-binding domain is a major determinant of differences in the effects of Ssa and Ssb proteins on [PSI(+)]. Surprisingly, overproduction of either chaperone increases loss of [PSI(+)] when Sup35 is simultaneously overproduced. Excess Ssa increases both the average size of prion polymers and the proportion of monomeric Sup35 protein. Both in vivo and in vitro experiments uncover direct physical interactions between Sup35 and Hsp70 proteins. The proposed model postulates that Ssa stimulates prion formation and polymer growth by stabilizing misfolded proteins, which serve as substrates for prion conversion. In the case of very large prion aggregates, further increase in size may lead to the loss of prion activity. In contrast, Ssb either stimulates refolding into nonprion conformation or targets misfolded proteins for degradation, in this way counteracting prion formation and propagation.
Figures





References
-
- Boorstein, W. R., and E. A. Craig, 1990. b Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem. 265: 18912–18921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

