RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases
- PMID: 15545645
- PMCID: PMC1449118
- DOI: 10.1534/genetics.104.035964
RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases
Abstract
The versatility of RNA-dependent RNA polymerases (RDRPs) in eukaryotic gene silencing is perhaps best illustrated in the kingdom Fungi. Biochemical and genetic studies of Schizosaccharomyces pombe and Neurospora crassa show that these types of enzymes are involved in a number of fundamental gene-silencing processes, including heterochromatin regulation and RNA silencing in S. pombe and meiotic silencing and RNA silencing in N. crassa. Here we show that Aspergillus nidulans, another model fungus, does not require an RDRP for inverted repeat transgene (IRT)-induced RNA silencing. However, RDRP requirements may vary within the Aspergillus genus as genomic analysis indicates that A. nidulans, but not A. fumigatus or A. oryzae, has lost a QDE-1 ortholog, an RDRP associated with RNA silencing in N. crassa. We also provide evidence suggesting that 5' --> 3' transitive RNA silencing is not a significant aspect of A. nidulans IRT-RNA silencing. These results indicate a lack of conserved kingdom-wide requirements for RDRPs in fungal RNA silencing.
Figures


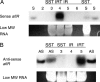


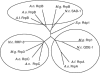
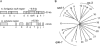
References
-
- Beclin, C., S. Boutet, P. Waterhouse and H. Vaucheret, 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688. - PubMed
-
- Bernstein, E., A. A. Caudy, S. M. Hammond and G. J. Hannon, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

