Genetic factors that regulate the attenuation of the general stress response of yeast
- PMID: 15545648
- PMCID: PMC1449543
- DOI: 10.1534/genetics.104.034603
Genetic factors that regulate the attenuation of the general stress response of yeast
Abstract
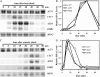
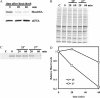
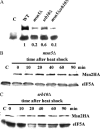
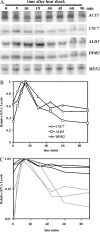
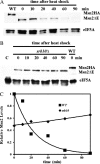
The general stress response of yeast involves the induction of approximately 200 genes in response to any one of several stresses. These genes are activated by Msn2 and repressed by the Srb10 kinase, a member of the mediator complex. Normally, Msn2 is exported from the nucleus, and Srb10 represses STRE gene expression. Under stress, Msn2 relocalizes to the nucleus and, with the relief of Srb10 repression, activates transcription. The stress response is rapid, but quickly attenuated. We show here that this attenuation is due to a nuclear-dependent degradation of Msn2. Msn2 rapidly disappeared from cells after heat or osmotic shock. This disappearance was not due to a change in MSN2 RNA levels, which remain constant during stress. Pulse-chase experiments confirmed the stress-dependent Msn2 degradation. The levels of Msn2 were significantly reduced in msn5 deletion cells that have been shown to constitutively retain Msn2 in the nucleus. The degradation was Srb10-dependent; Msn2 was not degraded in an srb10 deletion mutant. An Msn2 internal deletion mutant was insensitive to Srb10 repression, but was degraded by the Srb10-dependent mechanism. Thus, this mutation uncoupled Srb10 repression from degradation.
Figures


Similar articles
-
Role of Gal11, a component of the RNA polymerase II mediator in stress-induced hyperphosphorylation of Msn2 in Saccharomyces cerevisiae.Mol Microbiol. 2006 Oct;62(2):438-52. doi: 10.1111/j.1365-2958.2006.05363.x. Mol Microbiol. 2006. PMID: 17020582
-
Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase.Genes Dev. 2001 May 1;15(9):1078-92. doi: 10.1101/gad.867501. Genes Dev. 2001. PMID: 11331604 Free PMC article.
-
Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth.Mol Microbiol. 2000 Jan;35(2):397-406. doi: 10.1046/j.1365-2958.2000.01706.x. Mol Microbiol. 2000. PMID: 10652100
-
[Alternative ways of stress regulation in cells of Saccharomyces cerevisiae: transcriptional activators Msn2 and Msn4].Tsitologiia. 2009;51(3):271-8. Tsitologiia. 2009. PMID: 19435282 Review. Russian.
-
[Mechanisms of yeast resistance to environmental stress].Postepy Hig Med Dosw (Online). 2013 Apr 4;67:238-54. doi: 10.5604/17322693.1043394. Postepy Hig Med Dosw (Online). 2013. PMID: 23619223 Review. Polish.
Cited by
-
Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae.Sensors (Basel). 2010;10(6):6195-240. doi: 10.3390/s100606195. Epub 2010 Jun 21. Sensors (Basel). 2010. PMID: 22219709 Free PMC article. Review.
-
A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation.EMBO J. 2005 Dec 7;24(23):4115-23. doi: 10.1038/sj.emboj.7600871. Epub 2005 Nov 10. EMBO J. 2005. PMID: 16281053 Free PMC article.
-
The transcriptional activation region of Msn2p, in Saccharomyces cerevisiae, is regulated by stress but is insensitive to the cAMP signalling pathway.Mol Genet Genomics. 2006 Mar;275(3):277-87. doi: 10.1007/s00438-005-0017-4. Epub 2006 Feb 18. Mol Genet Genomics. 2006. PMID: 16489456
-
Global mRNA stability is not associated with levels of gene expression in Drosophila melanogaster but shows a negative correlation with codon bias.J Mol Evol. 2005 Sep;61(3):306-14. doi: 10.1007/s00239-004-0271-9. Epub 2005 Jul 21. J Mol Evol. 2005. PMID: 16044249
-
Transcriptomic and molecular genetic analysis of the cell wall salvage response of Aspergillus niger to the absence of galactofuranose synthesis.Cell Microbiol. 2016 Sep;18(9):1268-84. doi: 10.1111/cmi.12624. Epub 2016 Jul 29. Cell Microbiol. 2016. PMID: 27264789 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 2000 Current Protocols in Molecular Biology. John Wiley & Sons, New York.
-
- Beck, T., and M. N. Hall, 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

