Crystal structure of a group I intron splicing intermediate
- PMID: 15547134
- PMCID: PMC1370676
- DOI: 10.1261/rna.7140504
Crystal structure of a group I intron splicing intermediate
Abstract
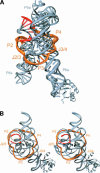
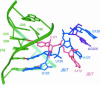
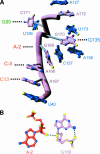
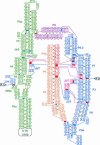
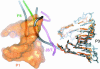
A recently reported crystal structure of an intact bacterial group I self-splicing intron in complex with both its exons provided the first molecular view into the mechanism of RNA splicing. This intron structure, which was trapped in the state prior to the exon ligation reaction, also reveals the architecture of a complex RNA fold. The majority of the intron is contained within three internally stacked, but sequence discontinuous, helical domains. Here the tertiary hydrogen bonding and stacking interactions between the domains, and the single-stranded joiner segments that bridge between them, are fully described. Features of the structure include: (1) A pseudoknot belt that circumscribes the molecule at its longitudinal midpoint; (2) two tetraloop-tetraloop receptor motifs at the peripheral edges of the structure; (3) an extensive minor groove triplex between the paired and joiner segments, P6-J6/6a and P3-J3/4, which provides the major interaction interface between the intron's two primary domains (P4-P6 and P3-P9.0); (4) a six-nucleotide J8/7 single stranded element that adopts a mu-shaped structure and twists through the active site, making critical contacts to all three helical domains; and (5) an extensive base stacking architecture that realizes 90% of all possible stacking interactions. The intron structure was validated by hydroxyl radical footprinting, where strong correlation was observed between experimental and predicted solvent accessibility. Models of the pre-first and pre-second steps of intron splicing are proposed with full-sized tRNA exons. They suggest that the tRNA undergoes substantial angular motion relative to the intron between the two steps of splicing.
Figures

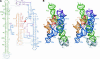












References
-
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. - PubMed
-
- Bass, B.L. and Cech, T.R. 1984. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: Implications for biological catalysis by RNA. Nature 308: 820–826. - PubMed
-
- ———. 1986. Ribozyme inhibitors: Deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry 25: 4473–4477. - PubMed
-
- Basu, S., Rambo, R.P., Strauss-Soukup, J., Cate, J.H., Ferre-D Amare, A.R., Strobel, S.A., and Doudna, J.A. 1998. A specific monovalent metal ion integral to the A-A platform of the RNA tetraloop receptor. Nat. Struct. Biol. 5: 986–992. - PubMed
-
- Been, M.D. 1994. Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem. Sci. 19: 251–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
