Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase
- PMID: 15547249
- PMCID: PMC534615
- DOI: 10.1093/nar/gkh923
Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase
Abstract
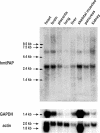
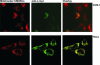
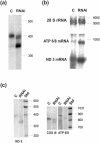
We report here on the identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Immunocytochemical experiments confirm that the enzyme indeed localizes to mitochondrial compartment. Inhibition of expression of the enzyme by RNA interference results in significant shortening of the poly(A) tails of the mitochondrial ND3, COX III and ATP 6/8 transcripts, suggesting that the investigated protein represents a bona fide mitochondrial poly(A) polymerase. This is in agreement with our sequencing data which show that poly(A) tails of several mitochondrial messengers are composed almost exclusively of adenosine residues. Moreover, the data presented here indicate that all analyzed mitochondrial transcripts with profoundly shortened poly(A) tails are relatively stable, which in turn argues against the direct role of long poly(A) extensions in the stabilization of human mitochondrial messengers.
Figures


References
-
- Manley J.L. and Proudfoot,N.J. (1994) RNA 3′ ends: formation and function–meeting review. Genes Dev., 8, 259–264. - PubMed
-
- Dreyfus M. and Regnier,P. (2002) The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell, 111, 611–613. - PubMed
-
- Regnier P. and Arraiano,C.M. (2000) Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays, 22, 235–244. - PubMed
-
- Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

