Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes
- PMID: 15547255
- PMCID: PMC529090
- DOI: 10.1128/JB.186.23.7847-7857.2004
Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes
Abstract
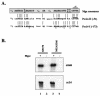
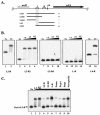
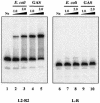
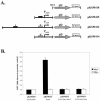
Streptococcus pyogenes (the group A streptococcus [GAS]) is a medically significant pathogen of humans, causing a range of diseases from pharyngitis to necrotizing fasciitis. Several important GAS virulence genes are under the control of a pleiotropic regulator called Mga, or the multiple gene regulator of GAS, including the gene encoding the streptococcal collagen-like protein, or sclA. Analysis of the genome sequence upstream of sclA revealed two potential Mga-binding sites with homology to the published Mga-binding element, which were called PsclA-I (distal) and PsclA-II (proximal) based on their location relative to a predicted start of transcription. Primer extension was used to confirm that the Mga-dependent transcriptional start site for sclA was located adjacent to the proximal PsclA-II binding site. By using overlapping PsclA promoter probes and purified Mga-His fusion protein, it was shown by electrophoretic mobility shift assays that, unlike other Mga-regulated promoters, Mga binds only to a distal DNA-binding site (PsclA-I). Binding of Mga to PsclA-I could be competed with cold probes corresponding to known Mga-regulated promoters (Pemm, PscpA, and Pmga) but not with a nonspecific probe or the proximal PsclA-II fragment. With the use of a plasmid-based green fluorescent protein transcriptional reporter system, the full-length PsclA was not sufficient to reproduce normal Mga-regulated activation. However, studies using a single-copy gusA transcriptional reporter system integrated at the native sclA chromosomal locus clearly demonstrated that the distal PsclA-I binding site is required for Mga regulation. Therefore, PsclA represents a new class of Mga-regulated promoters that requires a single distal binding site for activation.
Figures


References
-
- Andersson, G., K. McIver, L. O. Heden, and J. R. Scott. 1996. Complementation of divergent mga genes in group A streptococcus. Gene 175:77-81. - PubMed
-
- Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. - PubMed
-
- Chen, C., N. Bormann, and P. P. Cleary. 1993. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol. Gen. Genet. 241:685-693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

