Population structure and evolution of the Bacillus cereus group
- PMID: 15547268
- PMCID: PMC529064
- DOI: 10.1128/JB.186.23.7959-7970.2004
Population structure and evolution of the Bacillus cereus group
Abstract
Representative strains of the Bacillus cereus group of bacteria, including Bacillus anthracis (11 isolates), B. cereus (38 isolates), Bacillus mycoides (1 isolate), Bacillus thuringiensis (53 isolates from 17 serovars), and Bacillus weihenstephanensis (2 isolates) were assigned to 59 sequence types (STs) derived from the nucleotide sequences of seven alleles, glpF, gmk, ilvD, pta, pur, pycA, and tpi. Comparisons of the maximum likelihood (ML) tree of the concatenated sequences with individual gene trees showed more congruence than expected by chance, indicating a generally clonal structure to the population. The STs followed two major lines of descent. Clade 1 comprised B. anthracis strains, numerous B. cereus strains, and rare B. thuringiensis strains, while clade 2 included the majority of the B. thuringiensis strains together with some B. cereus strains. Other species were allocated to a third, heterogeneous clade. The ML trees and split decomposition analysis were used to assign STs to eight lineages within clades 1 and 2. These lineages were defined by bootstrap analysis and by a preponderance of fixed differences over shared polymorphisms among the STs. Lineages were named with reference to existing designations: Anthracis, Cereus I, Cereus II, Cereus III, Kurstaki, Sotto, Thuringiensis, and Tolworthi. Strains from some B. thuringiensis serovars were wholly or largely assigned to a single ST, for example, serovar aizawai isolates were assigned to ST-15, serovar kenyae isolates were assigned to ST-13, and serovar tolworthi isolates were assigned to ST-23, while other serovars, such as serovar canadensis, were genetically heterogeneous. We suggest a revision of the nomenclature in which the lineage and clone are recognized through name and ST designations in accordance with the clonal structure of the population.
Figures
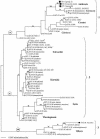
 , B. anthracis; ○, B. cereus; ▵, B. thuringiensis; ▪, B. mycoides; □, B. weihenstephanensis. All horizontal branch lengths were drawn to a scale of substitutions per site, and the tree was rooted at the midpoint for the purpose of clarity only. All bootstrap support values of >80% are shown next to the appropriate nodes. The 85% bootstrap value associated with clade 2 excludes the highly divergent ST-9 type.
, B. anthracis; ○, B. cereus; ▵, B. thuringiensis; ▪, B. mycoides; □, B. weihenstephanensis. All horizontal branch lengths were drawn to a scale of substitutions per site, and the tree was rooted at the midpoint for the purpose of clarity only. All bootstrap support values of >80% are shown next to the appropriate nodes. The 85% bootstrap value associated with clade 2 excludes the highly divergent ST-9 type.


Similar articles
-
Bacillus weihenstephanensis characteristics are present in Bacillus cereus and Bacillus mycoides strains.FEMS Microbiol Lett. 2013 Apr;341(2):127-37. doi: 10.1111/1574-6968.12106. Epub 2013 Mar 12. FEMS Microbiol Lett. 2013. PMID: 23413955
-
Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates.Appl Environ Microbiol. 2004 Feb;70(2):1068-80. doi: 10.1128/AEM.70.2.1068-1080.2004. Appl Environ Microbiol. 2004. PMID: 14766590 Free PMC article.
-
Multilocus sequence analysis of Bacillus thuringiensis serovars navarrensis, bolivia and vazensis and Bacillus weihenstephanensis reveals a common phylogeny.Antonie Van Leeuwenhoek. 2013 Jan;103(1):195-205. doi: 10.1007/s10482-012-9800-5. Epub 2012 Oct 17. Antonie Van Leeuwenhoek. 2013. PMID: 23073664
-
Sequence diversity of the Bacillus thuringiensis and B. cereus sensu lato flagellin (H antigen) protein: comparison with H serotype diversity.Appl Environ Microbiol. 2006 Jul;72(7):4653-62. doi: 10.1128/AEM.00328-06. Appl Environ Microbiol. 2006. PMID: 16820457 Free PMC article.
-
Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis.Can J Microbiol. 2007 Jun;53(6):673-87. doi: 10.1139/W07-029. Can J Microbiol. 2007. PMID: 17668027 Review.
Cited by
-
Signatures of selection in core and accessory genomes indicate different ecological drivers of diversification among Bacillus cereus clades.Mol Ecol. 2022 Jul;31(13):3584-3597. doi: 10.1111/mec.16490. Epub 2022 May 17. Mol Ecol. 2022. PMID: 35510788 Free PMC article.
-
Draft Genome Sequence of Bacillus cereus Strain BcFL2013, a Clinical Isolate Similar to G9241.Genome Announc. 2014 May 29;2(3):e00469-14. doi: 10.1128/genomeA.00469-14. Genome Announc. 2014. PMID: 24874674 Free PMC article.
-
Larvicidal, growth inhibitory and biochemical effects of soil bacterium, Pseudomonas sp. EN4 against Spodoptera litura (Fab.) (Lepidoptera: Noctuidae).BMC Microbiol. 2023 Apr 3;23(1):95. doi: 10.1186/s12866-023-02841-w. BMC Microbiol. 2023. PMID: 37013477 Free PMC article.
-
Investigating the genome diversity of B. cereus and evolutionary aspects of B. anthracis emergence.Genomics. 2011 Jul;98(1):26-39. doi: 10.1016/j.ygeno.2011.03.008. Epub 2011 Apr 5. Genomics. 2011. PMID: 21447378 Free PMC article.
-
Anthrax Toxin-Expressing Bacillus cereus Isolated from an Anthrax-Like Eschar.PLoS One. 2016 Jun 3;11(6):e0156987. doi: 10.1371/journal.pone.0156987. eCollection 2016. PLoS One. 2016. PMID: 27257909 Free PMC article.
References
-
- Ankarloo, J., D. A. Caugant, B. M. Hansen, A. Berg, A.-B. Kolstø, and A. Lovgren. 2000. Genome stability of Bacillus thuringiensis subsp. israelensis isolates. Curr. Microbiol. 40:51-56. - PubMed
-
- Ash, C., and M. D. Collins. 1992. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol. Lett. 73:75-80. - PubMed
-
- Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. - PubMed
-
- Carlson, C. R., T. Johansen, and A.-B. Kolstø. 1996. The chromosome map of Bacillus thuringiensis subsp. canadensis HD224 is highly similar to that of Bacillus cereus type strain ATCC 14579. FEMS Microbiol. Lett. 141:163-167. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

