Essentiality of the early transcript in the replication origin of the lactococcal prolate phage c2
- PMID: 15547273
- PMCID: PMC529073
- DOI: 10.1128/JB.186.23.8010-8017.2004
Essentiality of the early transcript in the replication origin of the lactococcal prolate phage c2
Abstract
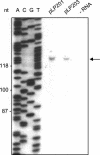
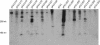
The genome of the prolate-headed lytic lactococcal bacteriophage c2 is organized into two divergently oriented blocks consisting of the early genes and the late genes. These blocks are separated by the noncoding origin of DNA replication. We examined the functional role of transcription of the origin in a plasmid model system. Deletion of the early promoter P(E)1 abolished origin function. Introduction of mutations into P(E)1 which did not eliminate promoter activity or replacement of P(E)1 with an unrelated but functional promoter did not abolish replication. The A-T-rich region upstream of P(E)1, which is conserved in prolate phages, was not required for plasmid replication. Replacement of the P(E)1 transcript template sequence with an unrelated sequence with a similar G+C content abolished replication, showing that the sequence encoding the transcript is essential for origin function. Truncated transcript and internal deletion constructs did not support replication except when the deletion was at the very 3' end of the DNA sequence coding for the transcript. The P(E)1 transcript could be detected for all replication-proficient constructs. Recloning in a plasmid vector allowed detection of P(E)1 transcripts from some fragments that did not support replication, indicating that stability of the transcript alone was not sufficient for replication. The data suggest that production of a transcript of a specific length and with a specific sequence or structure is essential for the function of the phage c2 origin in this model system.
Figures



Similar articles
-
Sequence diversity and functional conservation of the origin of replication in lactococcal prolate phages.Appl Environ Microbiol. 2003 Sep;69(9):5104-14. doi: 10.1128/AEM.69.9.5104-5114.2003. Appl Environ Microbiol. 2003. PMID: 12957892 Free PMC article.
-
Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1.Mol Microbiol. 1997 Oct;26(1):49-64. doi: 10.1046/j.1365-2958.1997.5491926.x. Mol Microbiol. 1997. PMID: 9383189
-
Examination of lactococcal bacteriophage c2 DNA replication using two-dimensional agarose gel electrophoresis.Gene. 2001 Oct 31;278(1-2):101-6. doi: 10.1016/s0378-1119(01)00702-8. Gene. 2001. PMID: 11707326
-
An origin of DNA replication from Lactococcus lactis bacteriophage c2.Appl Environ Microbiol. 1996 Apr;62(4):1452-3. doi: 10.1128/aem.62.4.1452-1453.1996. Appl Environ Microbiol. 1996. PMID: 8919811 Free PMC article.
-
The Lactococcus lactis plasmidome: much learnt, yet still lots to discover.FEMS Microbiol Rev. 2014 Sep;38(5):1066-88. doi: 10.1111/1574-6976.12074. Epub 2014 Jun 23. FEMS Microbiol Rev. 2014. PMID: 24861818 Review.
Cited by
-
LactococcusCeduovirus Phages Isolated from Industrial Dairy Plants-from Physiological to Genomic Analyses.Viruses. 2020 Mar 3;12(3):280. doi: 10.3390/v12030280. Viruses. 2020. PMID: 32138347 Free PMC article.
-
Isolation of lactococcal prolate phage-phage recombinants by an enrichment strategy reveals two novel host range determinants.J Bacteriol. 2005 May;187(9):3110-21. doi: 10.1128/JB.187.9.3110-3121.2005. J Bacteriol. 2005. PMID: 15838038 Free PMC article.
References
-
- Baker, T. A., and A. Kornberg. 1988. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell 55:113-123. - PubMed
-
- Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. - PubMed
-
- Callanan, M. J., P. W. O'Toole, M. W. Lubbers, and K. M. Polzin. 2001. Examination of lactococcal bacteriophage c2 DNA replication using two-dimensional agarose gel electrophoresis. Gene 278:101-106. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

