Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development
- PMID: 15547279
- PMCID: PMC529096
- DOI: 10.1128/JB.186.23.8066-8073.2004
Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development
Abstract
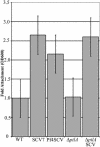
A current question in biofilm research is whether biofilm-specific genetic processes can lead to differentiation in physiology and function among biofilm cells. In Pseudomonas aeruginosa, phenotypic variants which exhibit a small-colony phenotype on agar media and a markedly accelerated pattern of biofilm development compared to that of the parental strain are often isolated from biofilms. We grew P. aeruginosa biofilms in glass flow cell reactors and observed that the emergence of small-colony variants (SCVs) in the effluent runoff from the biofilms correlated with the emergence of plaque-forming Pf1-like filamentous phage (designated Pf4) from the biofilm. Because several recent studies have shown that bacteriophage genes are among the most highly upregulated groups of genes during biofilm development, we investigated whether Pf4 plays a role in SCV formation during P. aeruginosa biofilm development. We carried out immunoelectron microscopy using anti-Pf4 antibodies and observed that SCV cells, but not parental-type cells, exhibited high densities of Pf4 filaments on the cell surface and that these filaments were often tightly interwoven into complex latticeworks surrounding the cells. Moreover, infection of P. aeruginosa planktonic cultures with Pf4 caused the emergence of SCVs within the culture. These SCVs exhibited enhanced attachment, accelerated biofilm development, and large regions of dead and lysed cells inside microcolonies in a manner identical to that of SCVs obtained from biofilms. We concluded that Pf4 can mediate phenotypic variation in P. aeruginosa biofilms. We also performed partial sequencing and analysis of the Pf4 replicative form and identified a number of open reading frames not previously recognized in the genome of P. aeruginosa, including a putative postsegregational killing operon.
Figures




References
-
- Agol, V. I. 1976. An aspect on the origin and evolution of viruses. Origins Life 7:119-132. - PubMed
-
- Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. - PubMed
-
- Bradley, D. E. 1973. The adsorption of the Pseudomonas aeruginosa filamentous bacteriophage Pf to its host. Can. J. Microbiol. 19:623-631. - PubMed
-
- Brinton, C. C., Jr. 1971. The properties of sex pili, the viral nature of “conjugal” genetic transfer systems, and some possible approaches to the control of bacterial drug resistance. Crit. Rev. Microbiol. 1:105-160. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

