Crystal structure of the PdxY Protein from Escherichia coli
- PMID: 15547280
- PMCID: PMC529075
- DOI: 10.1128/JB.186.23.8074-8082.2004
Crystal structure of the PdxY Protein from Escherichia coli
Abstract
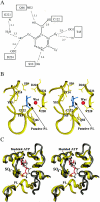
The crystal structure of Escherichia coli PdxY, the protein product of the pdxY gene, has been determined to a 2.2-A resolution. PdxY is a member of the ribokinase superfamily of enzymes and has sequence homology with pyridoxal kinases that phosphorylate pyridoxal at the C-5' hydroxyl. The protein is a homodimer with an active site on each monomer composed of residues that come exclusively from each respective subunit. The active site is filled with a density that fits that of pyridoxal. In monomer A, the ligand appears to be covalently attached to Cys122 as a thiohemiacetal, while in monomer B it is not covalently attached but appears to be partially present as pyridoxal 5'-phosphate. The presence of pyridoxal phosphate and pyridoxal as ligands was confirmed by the activation of aposerine hydroxymethyltransferase after release of the ligand by the denaturation of PdxY. The ligand, which appears to be covalently attached to Cys122, does not dissociate after denaturation of the protein. A detailed comparison (of functional properties, sequence homology, active site and ATP-binding-site residues, and active site flap types) of PdxY with other pyridoxal kinases as well as the ribokinase superfamily in general suggested that PdxY is a member of a new subclass of the ribokinase superfamily. The structure of PdxY also permitted an interpretation of work that was previously published about this enzyme.
Figures



References
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. - PubMed
-
- Cambillau, C., and E. Horjales. 1987. TOM: a Frodo subpackage for protein-ligand fitting with interactive energy minimization. J. Mol. Graph. 5:174-177.
-
- Campobasso, N., I. I. Mathews, T. P. Begley, and S. E. Ealick. 2000. Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry 39:7868-7877. - PubMed
-
- Cheng, G., E. M. Bennett, T. P. Begley, and S. E. Ealick. 2002. Crystal structure of 4-amino-5-hydroymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure 10:225-235. - PubMed
-
- Di Salvo, M. L., S. Hunt, and V. Schirch. 2004. Expression, purification and kinetic constants for human and Escherichia coli pyridoxal kinases. Protein Expr. Purif. 36:300-306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

