PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation
- PMID: 15548136
- PMCID: PMC1134783
- DOI: 10.1042/BJ20041638
PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation
Abstract
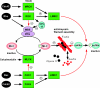
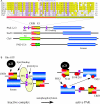
The Rho GTPases are a family of molecular switches that are critical regulators of signal transduction pathways in eukaryotic cells. They are known principally for their role in regulating the cytoskeleton, and do so by recruiting a variety of downstream effector proteins. Kinases form an important class of Rho effector, and part of the biological complexity brought about by switching on a single GTPase results from downstream phosphorylation cascades. Here we focus on our current understanding of the way in which different Rho-associated serine/threonine kinases, denoted PAK (p21-activated kinase), MLK (mixed-lineage kinase), ROK (Rho-kinase), MRCK (myotonin-related Cdc42-binding kinase), CRIK (citron kinase) and PKN (protein kinase novel), interact with and are regulated by their partner GTPases. All of these kinases have in common an ability to dimerize, and in most cases interact with a variety of other proteins that are important for their function. A diversity of known structures underpin the Rho GTPase-kinase interaction, but only in the case of PAK do we have a good molecular understanding of kinase regulation. The ability of Rho GTPases to co-ordinate spatial and temporal phosphorylation events explains in part their prominent role in eukaryotic cell biology.
Figures




References
-
- Madaule P., Axel R. A novel ras-related gene family. Cell. 1985;41:31–40. - PubMed
-
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. - PubMed
-
- Ridley A. J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. - PubMed
-
- Nobes C. D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. - PubMed
-
- Manser E., Leung T., Salihuddin H., Zhao Z. S., Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature (London) 1994;367:40–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

