A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness
- PMID: 15548738
- PMCID: PMC535875
- DOI: 10.1105/tpc.104.025973
A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness
Abstract
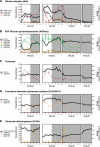
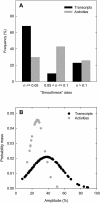
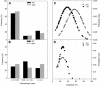
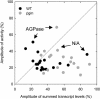
A platform has been developed to measure the activity of 23 enzymes that are involved in central carbon and nitrogen metabolism in Arabidopsis thaliana. Activities are assayed in optimized stopped assays and the product then determined using a suite of enzyme cycling assays. The platform requires inexpensive equipment, is organized in a modular manner to optimize logistics, calculates results automatically, combines high sensitivity with throughput, can be robotized, and has a throughput of three to four activities in 100 samples per person/day. Several of the assays, including those for sucrose phosphate synthase, ADP glucose pyrophosphorylase (AGPase), ferredoxin-dependent glutamate synthase, glycerokinase, and shikimate dehydrogenase, provide large advantages over previous approaches. This platform was used to analyze the diurnal changes of enzyme activities in wild-type Columbia-0 (Col-0) and the starchless plastid phosphoglucomutase (pgm) mutant, and in Col-0 during a prolongation of the night. The changes of enzyme activities were compared with the changes of transcript levels determined with the Affymetrix ATH1 array. Changes of transcript levels typically led to strongly damped changes of enzyme activity. There was no relation between the amplitudes of the diurnal changes of transcript and enzyme activity. The largest diurnal changes in activity were found for AGPase and nitrate reductase. Examination of the data and comparison with the literature indicated that these are mainly because of posttranslational regulation. The changes of enzyme activity are also strongly delayed, with the delay varying from enzyme to enzyme. It is proposed that enzyme activities provide a quasi-stable integration of regulation at several levels and provide useful data for the characterization and diagnosis of different physiological states. As an illustration, a decision tree constructed using data from Col-0 during diurnal changes and a prolonged dark treatment was used to show that, irrespective of the time of harvest during the diurnal cycle, the pgm mutant resembles a wild-type plant that has been exposed to a 3 d prolongation of the night.
Figures





References
-
- ap Rees, T., and Hill, S.A. (1994). Metabolic control analysis of plant metabolism. Plant Cell Environ. 17, 587–599.
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Ashour, M.B.A., Gee, S.J., and Hammock, B.D. (1987). Use of a 96-well microplate reader for measuring routine enzyme activities. Anal. Biochem. 166, 353–360. - PubMed
-
- Bergmeyer, H.U. (1987). Methods of Enzymatic Analysis. (Weinheim, Germany: VCH).
-
- Bernofsky, C., and Swan, M. (1973). Improved cycling assay for nicotinamide adenine-dinucleotide. Anal. Biochem. 53, 452–458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

