The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants
- PMID: 15548740
- PMCID: PMC535888
- DOI: 10.1105/tpc.104.026765
The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants
Abstract
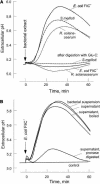
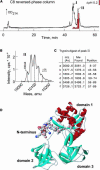
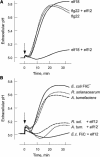
Innate immunity is based on the recognition of pathogen-associated molecular patterns (PAMPs). Here, we show that elongation factor Tu (EF-Tu), the most abundant bacterial protein, acts as a PAMP in Arabidopsis thaliana and other Brassicaceae. EF-Tu is highly conserved in all bacteria and is known to be N-acetylated in Escherichia coli. Arabidopsis plants specifically recognize the N terminus of the protein, and an N-acetylated peptide comprising the first 18 amino acids, termed elf18, is fully active as inducer of defense responses. The shorter peptide, elf12, comprising the acetyl group and the first 12 N-terminal amino acids, is inactive as elicitor but acts as a specific antagonist for EF-Tu-related elicitors. In leaves of Arabidopsis plants, elf18 induces an oxidative burst and biosynthesis of ethylene, and it triggers resistance to subsequent infection with pathogenic bacteria.
Figures





References
-
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gómez-Gómez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. - PubMed
-
- Baensch, M., Frank, R., and Kohl, J. (1998). Conservation of the amino-terminal epitope of elongation factor Tu in eubacteria and Archaea. Microbiology 144, 2241–2246. - PubMed
-
- Basse, C.W., Bock, K., and Boller, T. (1992). Elicitors and suppressors of the defense response in tomato cells. Purification and characterization of glycopeptide elicitors and glycan suppressors generated by enzymatic cleavage of yeast invertase. J. Biol. Chem. 267, 10258–10265. - PubMed
-
- Bauer, Z., Gómez-Gómez, L., Boller, T., and Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276, 45669–45676. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

