The stability of Seeman JX DNA topoisomers of paranemic crossover (PX) molecules as a function of crossover number
- PMID: 15550565
- PMCID: PMC534617
- DOI: 10.1093/nar/gkh931
The stability of Seeman JX DNA topoisomers of paranemic crossover (PX) molecules as a function of crossover number
Abstract
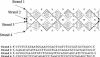
We use molecular dynamics simulations in explicit water and salt (Na+) to determine the effect of varying the number of crossover points on the structure and stability of the PX65 paranemic crossover DNA molecule and its JXM topoisomers (M denotes the number of missing crossover points), recently synthesized by the Seeman group at New York University. We find that PX65, with six crossover points, is the most stable, and that the stability decreases monotonically with the number of crossover points PX65 > JX1 > JX2 > JX3 > JX4, with 6, 5, 4, 3 and 2 crossover points, respectively. Thus, for PX65/JX1, the strain energy is approximately 3 kcal/mol/bp, while it is approximately 13 kcal/mol/bp for JX2, JX3 and JX4. Another measure of the stability is the change in the structure from the minimum energy structure to the equilibrium structure at 300 K, denoted as root-mean-square deviation in coordinates (CRMSD). We find that CRMSD is approximately 3.5 A for PX65, increases to 6 A for JX1 and increases to 10 A for JX2/JX3/JX4. As the number of crossover points decreases, the distance between the two double helical domains of the PX/JX molecules increases from approximately 20 A for PX65 to 23 A for JX4. This indicates that JX2, JX3 and JX4 are less likely to form, at least in with Na+. However, in all the cases, the two double helical domains have average helicoidal parameters similar to a typical B-DNA of similar length and base sequence.
Figures









References
-
- Seeman N.C. (2003) DNA in a material world. Nature, 421, 427–431. - PubMed
-
- Seeman N.C. (2003) Biochemistry and structural DNA nanotechnology: an evolving symbiotic relationship. Biochemistry, 42, 7259–7269. - PubMed
-
- Winfree E., Liu,F.R., Wenzler,L.A. and Seeman,N.C. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature, 394, 539–544. - PubMed
-
- Seeman N.C., Liu,F.R., Mao,C.D., Yang,X.P., Wenzler,L.A., Sha,R.J., Sun,W.Q., Shen,Z.Y., Li,X.J., Qi,J., Zhang,Y.W., Fu,T.J., Chen,J.H. and Winfree,E. (2000) Two dimensions and two states in DNA nanotechnology. J. Biomol. Struct. Dyn., 253–262. - PubMed
-
- Seeman N.C., Liu,F., Wenzler,L.A. and Winfree,E. (1999) Design and modification of two dimensional DNA arrays. Biophys. J., 76, A152–A152.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

