Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism
- PMID: 15555999
- PMCID: PMC3160629
- DOI: 10.1016/j.chembiol.2004.08.024
Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism
Abstract
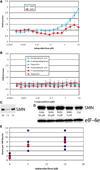
Most patients with the pediatric neurodegenerative disease spinal muscular atrophy have a homozygous deletion of the survival motor neuron 1 (SMN1) gene, but retain one or more copies of the closely related SMN2 gene. The SMN2 gene encodes the same protein (SMN) but produces it at a low efficiency compared with the SMN1 gene. We performed a high-throughput screen of approximately 47,000 compounds to identify those that increase production of an SMN2-luciferase reporter protein, but not an SMN1-luciferase reporter protein. Indoprofen, a nonsteroidal anti-inflammatory drug (NSAID) and cyclooxygenase (COX) inhibitor, selectively increased SMN2-luciferase reporter protein and endogenous SMN protein and caused a 5-fold increase in the number of nuclear gems in fibroblasts from SMA patients. No other NSAIDs or COX inhibitors tested exhibited this activity.
Figures
References
-
- Melki J, Lefebvre S, Burglen L, Burlet P, Clermont O, Millasseau P, Reboullet S, Benichou B, Zeviani M, Le Paslier D, et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994;264:1474–1477. - PubMed
-
- Melki J, Abdelhak S, Sheth P, Bachelot MF, Burlet P, Marcadet A, Aicardi J, Barois A, Carriere JP, Fardeau M, et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature. 1990;344:767–768. - PubMed
-
- Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;1:919–922. - PubMed
-
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

