A double-deletion method to quantifying incremental binding energies in proteins from experiment: example of a destabilizing hydrogen bonding pair
- PMID: 15556980
- PMCID: PMC1305133
- DOI: 10.1529/biophysj.104.050203
A double-deletion method to quantifying incremental binding energies in proteins from experiment: example of a destabilizing hydrogen bonding pair
Abstract
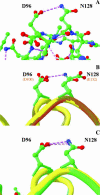
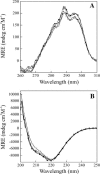
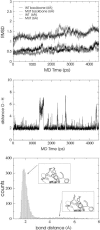
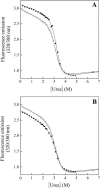
The contribution of a specific hydrogen bond in apoflavodoxin to protein stability is investigated by combining theory, experiment and simulation. Although hydrogen bonds are major determinants of protein structure and function, their contribution to protein stability is still unclear and widely debated. The best method so far devised to estimate the contribution of side-chain interactions to protein stability is double mutant cycle analysis, but the interaction energies so derived are not identical to incremental binding energies (the energies quantifying net contributions of two interacting groups to protein stability). Here we introduce double-deletion analysis of 'isolated' residue pairs as a means to precisely quantify incremental binding. The method is exemplified by studying a surface-exposed hydrogen bond in a model protein (Asp96/Asn128 in apoflavodoxin). Combined substitution of these residues by alanines slightly destabilizes the protein due to a decrease in hydrophobic surface burial. Subtraction of this effect, however, clearly indicates that the hydrogen-bonded groups in fact destabilize the native conformation. In addition, molecular dynamics simulations and classic double mutant cycle analysis explain quantitatively that, due to frustration, the hydrogen bond must form in the native structure because when the two groups get approximated upon folding their binding becomes favorable. We would like to remark that 1), this is the first time the contribution of a specific hydrogen bond to protein stability has been measured by experiment; and 2), more hydrogen bonds need to be analyzed to draw general conclusions on protein hydrogen bond energetics. To that end, the double-deletion method should be of help.
Figures


References
-
- Ben-Tal, N., D. Sitkoff, I. A. Topol, A.-S. Yang, S. Buro, and B. Ong. 1997. Free energy of amide hydrogen bond formation in vacuum, in water, and in liquid alkane solution. J. Phys. Chem. B. 101:450–457.
-
- Blaber, M., J. D. Lindstrom, N. Gassner, J. Xu, D. W. Heinz, and B. W. Matthews. 1993. Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala→Ser and Val→Thr substitutions in T4 lysozyme. Biochemistry. 32:11363–11373. - PubMed
-
- Branden, C., and J. Tooze. 1998. Introduction to Protein Structure. Garland Publishing, New York.
-
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. Status, S. Swaminathan, and M. J. Karplus. 1983. CHARMM: a program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem. 4:187–217.
-
- Carter, P. J., G. Winter, A. J. Wilkinson, and A. R. Fersht. 1984. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 38:835–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

