Evolution of new nonantibody proteins via iterative somatic hypermutation
- PMID: 15556995
- PMCID: PMC529417
- DOI: 10.1073/pnas.0407752101
Evolution of new nonantibody proteins via iterative somatic hypermutation
Abstract
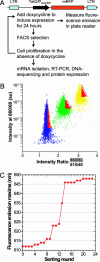
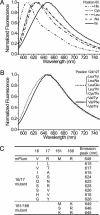
B lymphocytes use somatic hypermutation (SHM) to optimize immunoglobulins. Although SHM can rescue single point mutations deliberately introduced into nonimmunoglobulin genes, such experiments do not show whether SHM can efficiently evolve challenging novel phenotypes requiring multiple unforeseeable mutations in nonantibody proteins. We have now iterated SHM over 23 rounds of fluorescence-activated cell sorting to create monomeric red fluorescent proteins with increased photostability and far-red emissions (e.g., 649 nm), surpassing the best efforts of structure-based design. SHM offers a strategy to evolve nonantibody proteins with desirable properties for which a high-throughput selection or viable single-cell screen can be devised.
Figures

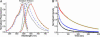

References
-
- Minshull, J. & Stemmer, W. P. C. (1999) Curr. Opin. Chem. Biol. 3, 284–290. - PubMed
-
- Petrounia, I. P. & Arnold, F. H. (2000) Curr. Opin. Biotechnol. 11, 325–330. - PubMed
-
- Greener, A., Callahan, M. & Jerpseth, B. (1997) Mol. Biotechnol. 7, 189–195. - PubMed
-
- Papavasiliou, F. N. & Schatz, D. G. (2002) Cell 109, Suppl., S35–S44. - PubMed
-
- Martin, A. & Scharff, M. D. (2002) Nat. Rev. Immunol. 2, 605–614. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

