Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis
- PMID: 15557095
- PMCID: PMC535826
- DOI: 10.1104/pp.104.051748
Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis
Erratum in
- Plant Physiol. 2005 Mar;137(3):1169. Hayashi, Kazuyuki [corrected to Hayashi, Kazunori]
Abstract
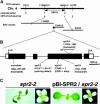
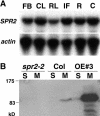
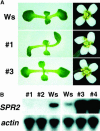
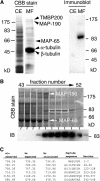
In diffusely growing plant cells, cortical microtubules play an important role in regulating the direction of cell expansion. Arabidopsis (Arabidopsis thaliana) spiral2 (spr2) mutant is defective in directional cell elongation and exhibits right-handed helical growth in longitudinally expanding organs such as root, hypocotyl, stem, petiole, and petal. The growth of spr2 roots is more sensitive to microtubule-interacting drugs than is wild-type root growth. The SPR2 gene encodes a plant-specific 94-kD protein containing HEAT-repeat motifs that are implicated in protein-protein interaction. When expressed constitutively, SPR2-green fluorescent protein fusion protein complemented the spr2 mutant phenotype and was localized to cortical microtubules as well as other mitotic microtubule arrays in transgenic plants. Recombinant SPR2 protein directly bound to taxol-stabilized microtubules in vitro. Furthermore, SPR2-specific antibody and mass spectrometry identified a tobacco (Nicotiana tabacum) SPR2 homolog in highly purified microtubule-associated protein fractions from tobacco BY-2 cell cultures. These results suggest that SPR2 is a novel microtubule-associated protein and is required for proper microtubule function involved in anisotropic growth.
Figures



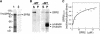
References
-
- Abe T, Thitamadee S, Hashimoto T (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol 45: 211–220 - PubMed
-
- An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153: 292–305
-
- Andrade MA, Bork P (1995) HEAT repeats in the Huntington's disease protein. Nat Genet 11: 115–116 - PubMed
-
- Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P (2001) Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1–18 - PubMed
-
- Andrade MA, Ponting CP, Gibson TJ, Bork P (2000) Homology-based method for identification of protein repeats using statistical significance estimates. J Mol Biol 298: 521–537 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

