First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157
- PMID: 15557626
- PMCID: PMC529153
- DOI: 10.1128/IAI.72.12.7030-7039.2004
First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157
Abstract
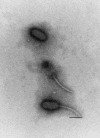
A bacteriophage encoding the Shiga toxin 2c variant (Stx2c) was isolated from the human Escherichia coli O157 strain CB2851 and shown to form lysogens on the E. coli K-12 laboratory strains C600 and MG1655. Production of Stx2c was found in the wild-type E. coli O157 strain and the K-12 lysogens and was inducible by growing bacteria in the presence of ciprofloxacin. Phage 2851 is the first reported viable bacteriophage which carries an stx(2c) gene. Electron micrographs of phage 2851 showed particles with elongated hexagonal heads and long flexible tails resembling phage lambda. Sequence analysis of an 8.4-kb region flanking the stx(2c) gene and other genetic elements revealed a mosaic gene structure, as found in other Stx phages. Phage 2851 showed lysis of E. coli K-12 strains lysogenic for Stx phages encoding Stx1 (H19), Stx2 (933W), Stx (7888), and Stx1c (6220) but showed superinfection immunity with phage lambda, presumably originating from the similarity of the cI repressor proteins of both phages. Apparently, phage 2851 integrates at a different chromosomal locus than Stx2 phage 933W and Stx1 phage H19 in E. coli, explaining why Stx2c is often found in combination with Stx1 or Stx2 in E. coli O157 strains. Diagnostic PCR was performed to determine gene sequences specific for phage 2851 in wild-type E. coli O157 strains producing Stx2c. The phage 2851 q and o genes were frequently detected in Stx2c-producing E. coli O157 strains, indicating that phages related to 2851 are associated with Stx2c production in strains of E. coli O157 that were isolated in different locations and time periods.
Figures


Similar articles
-
Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7.Infect Immun. 2008 Dec;76(12):5466-77. doi: 10.1128/IAI.00875-08. Epub 2008 Sep 29. Infect Immun. 2008. PMID: 18824528 Free PMC article.
-
Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle.Microbiology (Reading). 2004 Sep;150(Pt 9):2959-2971. doi: 10.1099/mic.0.27188-0. Microbiology (Reading). 2004. PMID: 15347754
-
Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei.Infect Immun. 2001 Dec;69(12):7588-95. doi: 10.1128/IAI.69.12.7588-7595.2001. Infect Immun. 2001. PMID: 11705937 Free PMC article.
-
Occurrence of Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli in the environment.Environ Sci Technol. 2006 Dec 1;40(23):7141-9. doi: 10.1021/es060927k. Environ Sci Technol. 2006. PMID: 17180960 Review.
-
Bacteriophage lambda: alive and well and still doing its thing.Curr Opin Microbiol. 2001 Apr;4(2):201-7. doi: 10.1016/s1369-5274(00)00189-2. Curr Opin Microbiol. 2001. PMID: 11282477 Review.
Cited by
-
Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks.Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4868-73. doi: 10.1073/pnas.0710834105. Epub 2008 Mar 10. Proc Natl Acad Sci U S A. 2008. PMID: 18332430 Free PMC article.
-
Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM.Appl Environ Microbiol. 2006 Mar;72(3):1900-9. doi: 10.1128/AEM.72.3.1900-1909.2006. Appl Environ Microbiol. 2006. PMID: 16517637 Free PMC article.
-
A single-step purification and molecular characterization of functional Shiga toxin 2 variants from pathogenic Escherichia coli.Toxins (Basel). 2012 Jul;4(7):487-504. doi: 10.3390/toxins4070487. Epub 2012 Jun 25. Toxins (Basel). 2012. PMID: 22852065 Free PMC article.
-
Diversity of Shiga toxin transducing phages in Escherichia coli O145:H28 and the different Shiga toxin 2 production levels associated with short- or long-tailed phages.Front Microbiol. 2024 Aug 6;15:1453887. doi: 10.3389/fmicb.2024.1453887. eCollection 2024. Front Microbiol. 2024. PMID: 39165568 Free PMC article.
-
Epidemiology of Shiga Toxin-Producing Escherichia coli O157 in the Province of Alberta, Canada, 2009-2016.Toxins (Basel). 2019 Oct 22;11(10):613. doi: 10.3390/toxins11100613. Toxins (Basel). 2019. PMID: 31652648 Free PMC article.
References
-
- Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
-
- Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. - PubMed
-
- Beutin, L., S. Kaulfuss, T. Cheasty, B. Brandenburg, S. Zimmermann, K. Gleier, G. A. Willshaw, and H. R. Smith. 2002. Characteristics and association with disease of two major subclones of Shiga toxin (Verocytotoxin)-producing strains of Escherichia coli (STEC) O157 that are present among isolates from patients in Germany. Diagn. Microbiol. Infect. 44:337-346. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

