Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans
- PMID: 15557647
- PMCID: PMC529107
- DOI: 10.1128/IAI.72.12.7220-7230.2004
Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans
Abstract
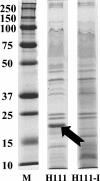
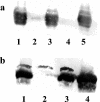
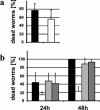
Burkholderia cenocepacia H111, which was isolated from a cystic fibrosis patient, employs a quorum-sensing (QS) system, encoded by cep, to control the expression of virulence factors as well as the formation of biofilms. The QS system is thought to ensure that pathogenic traits are expressed only when the bacterial population density is high enough to overwhelm the host before it is able to mount an efficient response. While the wild-type strain effectively kills the nematode Caenorhabditis elegans, the pathogenicity of mutants with defective quorum sensing is attenuated. To date, very little is known about the cep-regulated virulence factors required for nematode killing. Here we report the identification of a cep-regulated gene, whose predicted amino acid sequence is highly similar to the QS-regulated protein AidA of the plant pathogen Ralstonia solanacearum. By use of polyclonal antibodies directed against AidA, it is demonstrated that the protein is expressed in the late-exponential phase and accumulates during growth arrest. We show that B. cenocepacia H111 AidA is essential for slow killing of C. elegans but has little effect on fast killing, suggesting that the protein plays a role in the accumulation of the strain in the nematode gut. Thus, AidA appears to be required for establishing an infection-like process rather than acting as a toxin. Furthermore, evidence is provided that AidA is produced not only by B. cenocepacia but also by many other strains of the Burkholderia cepacia complex.
Figures



References
-
- Aballay, A., and F. M. Ausubel. 2002. Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5:97-101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

