Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center
- PMID: 15561858
- PMCID: PMC529220
- DOI: 10.1128/AAC.48.12.4793-4799.2004
Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center
Abstract
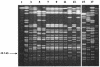
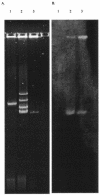
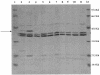
From April 2000 to April 2001, 24 patients in intensive care units at Tisch Hospital, New York, N.Y., were infected or colonized by carbapenem-resistant Klebsiella pneumoniae. Pulsed-field gel electrophoresis identified a predominant outbreak strain, but other resistant strains were also recovered. Three representatives of the outbreak strain from separate patients were studied in detail. All were resistant or had reduced susceptibility to imipenem, meropenem, ceftazidime, piperacillin-tazobactam, and gentamicin but remained fully susceptible to tetracycline. PCR amplified a blaKPC allele encoding a novel variant, KPC-3, with a His(272)-->Tyr substitution not found in KPC-2; other carbapenemase genes were absent. In the outbreak strain, KPC-3 was encoded by a 75-kb plasmid, which was transferred in vitro by electroporation and conjugation. The isolates lacked the OmpK35 porin but expressed OmpK36, implying reduced permeability as a cofactor in resistance. This is the third KPC carbapenem-hydrolyzing beta-lactamase variant to have been reported in members of the Enterobacteriaceae, with others reported from the East Coast of the United States. Although producers of these enzymes remain rare, the progress of this enzyme group merits monitoring.
Figures

References
-
- Anonymous. 2000. National Nosocomial Infection Surveillance (NNIS) System Report, data summary from January 1992-April 2000, issued June 2000. Am. J. Infect. Control 28:429-448. - PubMed
-
- Bou, G., G. Cerveró, M. A. Domínguez, C. Quereda, and J. Martínez-Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. - PMC - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55-60. - PubMed
-
- Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

