Genomewide linkage searches for Mendelian disease loci can be efficiently conducted using high-density SNP genotyping arrays
- PMID: 15561999
- PMCID: PMC534642
- DOI: 10.1093/nar/gnh163
Genomewide linkage searches for Mendelian disease loci can be efficiently conducted using high-density SNP genotyping arrays
Abstract
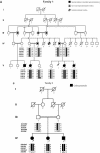
Genomewide linkage searches aimed at identifying disease susceptibility loci are generally conducted using 300-400 microsatellite markers. Genotyping bi-allelic single nucleotide polymorphisms (SNPs) provides an alternative strategy. The availability of dense SNP maps coupled with recent technological developments in highly paralleled SNP genotyping makes it practical to now consider the use of these markers for whole-genome genetic linkage analyses. Here, we report the findings from three successful genomewide linkage analyses of families segregating autosomal recessively inherited neonatal diabetes, craniosynostosis and dominantly inherited renal dysplasia using the Affymetrix 10K SNP array. A single locus was identified for each disease state, two of which are novel. The performance of the SNP array, both in terms of efficiency and precision, indicates that such platforms will become the dominant technology for performing genomewide linkage searches.
Figures




Similar articles
-
[Advances in high-density whole genome-wide single nucleotide polymorphism array in cancer research].Ai Zheng. 2006 Nov;25(11):1454-8. Ai Zheng. 2006. PMID: 17094921 Review. Chinese.
-
Design of tag SNP whole genome genotyping arrays.Methods Mol Biol. 2009;529:51-61. doi: 10.1007/978-1-59745-538-1_4. Methods Mol Biol. 2009. PMID: 19381970
-
Single nucleotide polymorphism mapping array assay.Methods Mol Biol. 2007;396:295-314. doi: 10.1007/978-1-59745-515-2_19. Methods Mol Biol. 2007. PMID: 18025700
-
Genomewide high-density SNP linkage analysis of non-BRCA1/2 breast cancer families identifies various candidate regions and has greater power than microsatellite studies.BMC Genomics. 2007 Aug 30;8:299. doi: 10.1186/1471-2164-8-299. BMC Genomics. 2007. PMID: 17760956 Free PMC article.
-
Applications of whole-genome high-density SNP genotyping.Expert Rev Mol Diagn. 2005 Mar;5(2):159-70. doi: 10.1586/14737159.5.2.159. Expert Rev Mol Diagn. 2005. PMID: 15833046 Review.
Cited by
-
Biallelic mutation of protocadherin-21 (PCDH21) causes retinal degeneration in humans.Mol Vis. 2010 Jan 15;16:46-52. Mol Vis. 2010. PMID: 20087419 Free PMC article.
-
A high-density SNP genomewide linkage scan for chronic lymphocytic leukemia-susceptibility loci.Am J Hum Genet. 2005 Sep;77(3):420-9. doi: 10.1086/444472. Epub 2005 Aug 2. Am J Hum Genet. 2005. PMID: 16080117 Free PMC article.
-
Locating potentially lethal genes using the abnormal distributions of genotypes.Sci Rep. 2019 Jul 22;9(1):10543. doi: 10.1038/s41598-019-47076-w. Sci Rep. 2019. PMID: 31332212 Free PMC article.
-
The Opdc missense mutation of Pax2 has a milder than loss-of-function phenotype.Hum Mol Genet. 2011 Jan 15;20(2):223-34. doi: 10.1093/hmg/ddq457. Epub 2010 Oct 13. Hum Mol Genet. 2011. PMID: 20943750 Free PMC article.
-
SNPs Array Karyotyping in Non-Hodgkin Lymphoma.Microarrays (Basel). 2015 Nov 12;4(4):551-69. doi: 10.3390/microarrays4040551. Microarrays (Basel). 2015. PMID: 27600240 Free PMC article. Review.
References
-
- Weissenbach J., Gyapay.G., Dib,C., Vignal,A., Morissette,J., Millasseau,P., Vaysseix,G. and Lathrop,M. (1992) A second-generation linkage map of the human genome. Nature, 359, 794–801. - PubMed
-
- Sheffield V.C., Weber,J.L., Buetow,K.H., Murray,J.C., Even,D.A., Wiles,K., Gastier,J.M., Pulido,J.C., Yandava,C. and Sunden,S.L. (1995) A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genomewide linkage maps. Hum. Mol. Genet., 4, 1837–1844. - PubMed
-
- Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. - PubMed
-
- Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

