The preferential binding of histone H1 to DNA scaffold-associated regions is determined by its C-terminal domain
- PMID: 15562002
- PMCID: PMC534626
- DOI: 10.1093/nar/gkh945
The preferential binding of histone H1 to DNA scaffold-associated regions is determined by its C-terminal domain
Abstract
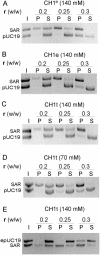
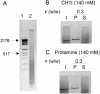
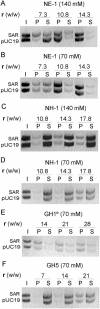
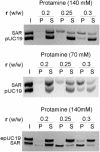
Histone H1 preferentially binds and aggregates scaffold-associated regions (SARs) via the numerous homopolymeric oligo(dA).oligo(dT) tracts present within these sequences. Here we show that the mammalian somatic subtypes H1a,b,c,d,e and H1 degrees and the male germline-specific subtype H1t, all preferentially bind to the Drosophila histone SAR. Experiments with the isolated domains show that whilst the C-terminal domain maintains strong and preferential binding, the N-terminal and globular domains show weak binding and poor specificity for the SAR. The preferential binding of SAR by the H1 molecule thus appears to be determined by its highly basic C-terminal domain. Salmine, a typical fish protamine, which could have its evolutionary origin in histone H1, also shows preferential binding to the SAR. The interaction of distamycin, a minor groove binder with high affinity for homopolymeric oligo(dA).oligo(dT) tracts, abolishes preferential binding of the C-terminal domain of histone H1 and protamine to the SAR, suggesting the involvement of the DNA minor groove in the interaction.
Figures


Similar articles
-
Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts.J Mol Biol. 1989 Dec 5;210(3):587-99. doi: 10.1016/0022-2836(89)90134-4. J Mol Biol. 1989. PMID: 2614835
-
Highly preferential nucleation of histone H1 assembly on scaffold-associated regions.J Mol Biol. 1989 Dec 5;210(3):573-85. doi: 10.1016/0022-2836(89)90133-2. J Mol Biol. 1989. PMID: 2614834
-
Preferential condensation of SAR-DNA by histone H1 and its SPKK containing octapeptide repeat motif.FEBS Lett. 1997 Jan 3;400(2):193-6. doi: 10.1016/s0014-5793(96)01393-2. FEBS Lett. 1997. PMID: 9001396
-
Post-translational modifications of the intrinsically disordered terminal domains of histone H1: effects on secondary structure and chromatin dynamics.Chromosoma. 2017 Feb;126(1):83-91. doi: 10.1007/s00412-016-0591-8. Epub 2016 Apr 21. Chromosoma. 2017. PMID: 27098855 Review.
-
Origin of H1 linker histones.FASEB J. 2001 Jan;15(1):34-42. doi: 10.1096/fj.00-0237rev. FASEB J. 2001. PMID: 11149891 Review.
Cited by
-
Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1.Biophys J. 2007 Sep 15;93(6):2170-7. doi: 10.1529/biophysj.107.104513. Epub 2007 May 18. Biophys J. 2007. PMID: 17513371 Free PMC article.
-
Functional equivalence of HMGA- and histone H1-like domains in a bacterial transcriptional factor.Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13546-51. doi: 10.1073/pnas.0902233106. Epub 2009 Jul 28. Proc Natl Acad Sci U S A. 2009. PMID: 19666574 Free PMC article.
-
Secondary structure of protamine in sperm nuclei: an infrared spectroscopy study.BMC Struct Biol. 2011 Mar 24;11:14. doi: 10.1186/1472-6807-11-14. BMC Struct Biol. 2011. PMID: 21435240 Free PMC article.
-
Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin.BMC Biol. 2007 May 11;5:22. doi: 10.1186/1741-7007-5-22. BMC Biol. 2007. PMID: 17498293 Free PMC article.
-
Proteasome-dependent degradation of histone H1 subtypes is mediated by its C-terminal domain.Protein Sci. 2024 May;33(5):e4970. doi: 10.1002/pro.4970. Protein Sci. 2024. PMID: 38591484 Free PMC article.
References
-
- Zlatanova J. and van Holde,K. (1992) Histone H1 and transcription: still an enigma? J. Cell Sci., 103, 889–895. - PubMed
-
- Bouvet L., Dimitrov,S. and Wolffe,A.P.K. (1994) Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev., 8, 1147–1159. - PubMed
-
- Khochbin S. and Wolffe,A.P. (1994) Developmentally regulated expression of linker-histone variants in vertebrates. Eur. J. Biochem., 225, 501–510. - PubMed
-
- Shen X. and Gorovsky,M.A. (1996) Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell, 86, 475–483. - PubMed
-
- Wolffe A.P., Khochbin,P.S. and Dimitrov,S. (1997) What do linker histones do in chromatin? BioEssays, 19, 249–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

