Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily
- PMID: 15562004
- PMCID: PMC534630
- DOI: 10.1093/nar/gkh951
Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily
Abstract
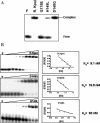
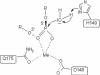
The restriction endonuclease (REase) R.KpnI is an orthodox Type IIP enzyme, which binds to DNA in the absence of metal ions and cleaves the DNA sequence 5'-GGTAC--C-3' in the presence of Mg2+ as shown generating 3' four base overhangs. Bioinformatics analysis reveals that R.KpnI contains a betabetaalpha-Me-finger fold, which is characteristic of many HNH-superfamily endonucleases, including homing endonuclease I-HmuI, structure-specific T4 endonuclease VII, colicin E9, sequence non-specific Serratia nuclease and sequence-specific homing endonuclease I-PpoI. According to our homology model of R.KpnI, D148, H149 and Q175 correspond to the critical D, H and N or H residues of the HNH nucleases. Substitutions of these three conserved residues lead to the loss of the DNA cleavage activity by R.KpnI, confirming their importance. The mutant Q175E fails to bind DNA at the standard conditions, although the DNA binding and cleavage can be rescued at pH 6.0, indicating a role for Q175 in DNA binding and cleavage. Our study provides the first experimental evidence for a Type IIP REase that does not belong to the PD...D/EXK superfamily of nucleases, instead is a member of the HNH superfamily.
Figures


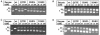

Similar articles
-
Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily.BMC Struct Biol. 2007 Jul 12;7:48. doi: 10.1186/1472-6807-7-48. BMC Struct Biol. 2007. PMID: 17626614 Free PMC article.
-
Generation of a manganese specific restriction endonuclease with nicking activity.Biochemistry. 2010 Sep 28;49(38):8425-33. doi: 10.1021/bi101035k. Biochemistry. 2010. PMID: 20734974
-
Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease.Proteins. 2006 Dec 1;65(4):867-76. doi: 10.1002/prot.21156. Proteins. 2006. PMID: 17029241
-
Type II restriction endonucleases: structure and mechanism.Cell Mol Life Sci. 2005 Mar;62(6):685-707. doi: 10.1007/s00018-004-4513-1. Cell Mol Life Sci. 2005. PMID: 15770420 Free PMC article. Review.
-
Catalytic mechanisms of restriction and homing endonucleases.Biochemistry. 2002 Nov 26;41(47):13851-60. doi: 10.1021/bi020467h. Biochemistry. 2002. PMID: 12437341 Review.
Cited by
-
Tetrameric structure of the restriction DNA glycosylase R.PabI in complex with nonspecific double-stranded DNA.Sci Rep. 2016 Oct 12;6:35197. doi: 10.1038/srep35197. Sci Rep. 2016. PMID: 27731370 Free PMC article.
-
Recognition and cleavage of 5-methylcytosine DNA by bacterial SRA-HNH proteins.Nucleic Acids Res. 2015 Jan;43(2):1147-59. doi: 10.1093/nar/gku1376. Epub 2015 Jan 6. Nucleic Acids Res. 2015. PMID: 25564526 Free PMC article.
-
Members of the RAD52 Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in Saccharomyces cerevisiae.PLoS Genet. 2015 Nov 5;11(11):e1005664. doi: 10.1371/journal.pgen.1005664. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26540255 Free PMC article.
-
Crystal structure and DNA cleavage mechanism of the restriction DNA glycosylase R.CcoLI from Campylobacter coli.Sci Rep. 2021 Jan 13;11(1):859. doi: 10.1038/s41598-020-79537-y. Sci Rep. 2021. PMID: 33441677 Free PMC article.
-
Inference of relationships in the 'twilight zone' of homology using a combination of bioinformatics and site-directed mutagenesis: a case study of restriction endonucleases Bsp6I and PvuII.Nucleic Acids Res. 2005 Jan 31;33(2):661-71. doi: 10.1093/nar/gki213. Print 2005. Nucleic Acids Res. 2005. PMID: 15684412 Free PMC article.
References
-
- Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type II restriction endonucleases. Eur. J. Biochem., 246, 1–22. - PubMed
-
- Kovall R.A. and Mathews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships, Curr. Opin. Chem. Biol., 3, 578–583. - PubMed
-
- Roberts R.J. and Halford,S.E. (1993) Type II restriction enzymes. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor, NY, pp 35–88.
-
- Grabowski G., Jeltsch,A., Wolfes,H., Maass,G. and Alves,J. (1995) Site-directed mutagenesis in the catalytic center of the restriction endonuclease. Gene, 157, 113–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

