The relative flexibility of B-DNA and A-RNA duplexes: database analysis
- PMID: 15562006
- PMCID: PMC534633
- DOI: 10.1093/nar/gkh954
The relative flexibility of B-DNA and A-RNA duplexes: database analysis
Abstract
An extensive analysis of structural databases is carried out to investigate the relative flexibility of B-DNA and A-RNA duplexes in crystal form. Our results show that the general anisotropic concept of flexibility is not very useful to compare the deformability of B-DNA and A-RNA duplexes, since the flexibility patterns of B-DNA and A-RNA are quite different. In other words, 'flexibility' is a dangerous word for describing macromolecules, unless it is clearly defined. A few soft essential movements explain most of the natural flexibility of A-RNA, whereas many are necessary for B-DNA. Essential movements occurring in naked B-DNAs are identical to those necessary to deform DNA in DNA-protein complexes, which suggest that evolution has designed DNA-protein complexes so that B-DNA is deformed according to its natural tendency. DNA is generally more flexible, but for some distortions A-RNA is easier to deform. Local stiffness constants obtained for naked B-DNAs and DNA complexes are very close, demonstrating that global distortions in DNA necessary for binding to proteins are the result of the addition of small concerted deformations at the base-pair level. Finally, it is worth noting that in general the picture of the relative deformability of A-RNA and DNA derived from database analysis agrees very well with that derived from state of the art molecular dynamics (MD) simulations.
Figures

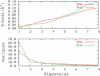
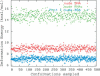
References
-
- Bloomfield V.A., Tinoco,I., Hearst,J.E. and Crothers,D.M. (2000) Nucleic Acids: Structures, Properties, and Functions. University Science Books, Sausalito, CA.
-
- Gait M.J. and Blackburn,G.M. (1990) Nucleic Acids in Chemistry and Biology. IRL Press, Oxford University Press, Oxford.
-
- Saenger W. (1984) Principles of Nucleic Acid Structure. Springer, NY.
-
- Cluzel P., Lebrun,A., Heller,C., Lavery,R., Viovy,J.L., Chatenay,D. and Caron,F. (1996) DNA: an extensible molecule. Science, 271, 792–794. - PubMed

