A new mixture model approach to analyzing allelic-loss data using Bayes factors
- PMID: 15563371
- PMCID: PMC544187
- DOI: 10.1186/1471-2105-5-182
A new mixture model approach to analyzing allelic-loss data using Bayes factors
Abstract
Background: Allelic-loss studies record data on the loss of genetic material in tumor tissue relative to normal tissue at various loci along the genome. As the deletion of a tumor suppressor gene can lead to tumor development, one objective of these studies is to determine which, if any, chromosome arms harbor tumor suppressor genes.
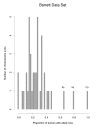
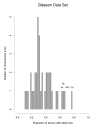
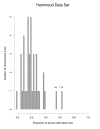
Results: We propose a large class of mixture models for describing the data, and we suggest using Bayes factors to select a reasonable model from the class in order to classify the chromosome arms. Bayes factors are especially useful in the case of testing that the number of components in a mixture model is n0 versus n1. In these cases, frequentist test statistics based on the likelihood ratio statistic have unknown distributions and are therefore not applicable. Our simulation study shows that Bayes factors favor the right model most of the time when tumor suppressor genes are present. When no tumor suppressor genes are present and background allelic-loss varies, the Bayes factors are often inconclusive, although this results in a markedly reduced false-positive rate compared to that of standard frequentist approaches. Application of our methods to three data sets of esophageal adenocarcinomas yields interesting differences from those results previously published.
Conclusions: Our results indicate that Bayes factors are useful for analyzing allelic-loss data.
Figures
Similar articles
-
Allelic deletion in 11p15 is a common occurrence in esophageal and gastric adenocarcinoma.Cancer. 1998 Jul 15;83(2):232-9. Cancer. 1998. PMID: 9669804
-
Loss of heterozygosity involves multiple tumor suppressor genes in human esophageal cancers.Cancer Res. 1992 Dec 1;52(23):6525-30. Cancer Res. 1992. PMID: 1423299
-
Inferring the location and effect of tumor suppressor genes by instability-selection modeling of allelic-loss data.Biometrics. 2000 Dec;56(4):1088-97. doi: 10.1111/j.0006-341x.2000.01088.x. Biometrics. 2000. PMID: 11129465
-
Inferring the location of tumor suppressor genes by modeling frequency of allelic loss.Biometrics. 2007 Mar;63(1):33-40. doi: 10.1111/j.1541-0420.2006.00636.x. Biometrics. 2007. PMID: 17447927
-
Identifying multiple changepoints in heterogeneous binary data with an application to molecular genetics.Biostatistics. 2004 Oct;5(4):515-29. doi: 10.1093/biostatistics/kxh005. Biostatistics. 2004. PMID: 15475416
Cited by
-
Identification of pharmacogenetic markers in smoking cessation therapy.Am J Med Genet B Neuropsychiatr Genet. 2008 Sep 5;147B(6):712-9. doi: 10.1002/ajmg.b.30669. Am J Med Genet B Neuropsychiatr Genet. 2008. PMID: 18165968 Free PMC article. Clinical Trial.
References
-
- Barrett MT, Galipeau PC, Sanchez CA, Emond MJ, Reid BJ. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene. 1996;12:1873–1878. - PubMed
-
- Hammoud ZT, Kaleem Z, Cooper JD, Sundaresan RS, Patterson GA, Goodfellow PJ. Allelotype analysis of esophageal adenocarcinomas: evidence for the involvement of sequences on the long arm of chromosome 4. Cancer Research. 1996;56:4499–4502. - PubMed
-
- Fearon ER. Tumor suppressor genes. The Genetic Basis of Human Cancer. 1998;7:145.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials