The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
- PMID: 15563598
- PMCID: PMC535379
- DOI: 10.1073/pnas.0406475101
The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
Abstract
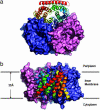
Ammonium is one of the most important nitrogen sources for bacteria, fungi, and plants, but it is toxic to animals. The ammonium transport proteins (methylamine permeases/ammonium transporters/rhesus) are present in all domains of life; however, functional studies with members of this family have yielded controversial results with respect to the chemical identity (NH(4)(+) or NH(3)) of the transported species. We have solved the structure of wild-type AmtB from Escherichia coli in two crystal forms at 1.8- and 2.1-A resolution, respectively. Substrate transport occurs through a narrow mainly hydrophobic pore located at the center of each monomer of the trimeric AmtB. At the periplasmic entry, a binding site for NH(4)(+) is observed. Two phenylalanine side chains (F107 and F215) block access into the pore from the periplasmic side. Further into the pore, the side chains of two highly conserved histidine residues (H168 and H318) bridged by a H-bond lie adjacent, with their edges pointing into the cavity. These histidine residues may facilitate the deprotonation of an ammonium ion entering the pore. Adiabatic free energy calculations support the hypothesis that an electrostatic barrier between H168 and H318 hinders the permeation of cations but not that of the uncharged NH(3.) The structural data and energetic considerations strongly indicate that the methylamine permeases/ammonium transporters/rhesus proteins are ammonia gas channels. Interestingly, at the cytoplasmic exit of the pore, two different conformational states are observed that might be related to the inactivation mechanism by its regulatory partner.
Figures



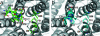
Similar articles
-
Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.Proc Natl Acad Sci U S A. 2008 Apr 1;105(13):5040-5. doi: 10.1073/pnas.0711742105. Epub 2008 Mar 24. Proc Natl Acad Sci U S A. 2008. PMID: 18362341 Free PMC article.
-
Structural and functional insights into the AmtB/Mep/Rh protein family.Transfus Clin Biol. 2006 Mar-Apr;13(1-2):65-9. doi: 10.1016/j.tracli.2006.02.014. Epub 2006 Mar 24. Transfus Clin Biol. 2006. PMID: 16564194 Review.
-
Ammonium recruitment and ammonia transport by E. coli ammonia channel AmtB.Biophys J. 2006 Dec 15;91(12):4401-12. doi: 10.1529/biophysj.106.089714. Epub 2006 Sep 29. Biophys J. 2006. PMID: 17012311 Free PMC article.
-
The pivotal twin histidines and aromatic triad of the Escherichia coli ammonium channel AmtB can be replaced.Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):13270-4. doi: 10.1073/pnas.1108451108. Epub 2011 Jul 20. Proc Natl Acad Sci U S A. 2011. PMID: 21775672 Free PMC article.
-
Amt/MEP/Rh proteins conduct ammonia.Pflugers Arch. 2006 Mar;451(6):701-7. doi: 10.1007/s00424-005-1511-6. Epub 2005 Nov 5. Pflugers Arch. 2006. PMID: 16273393 Review.
Cited by
-
Deinococcus radiodurans UWO298 Dependence on Background Radiation for Optimal Growth.Front Genet. 2021 May 6;12:644292. doi: 10.3389/fgene.2021.644292. eCollection 2021. Front Genet. 2021. PMID: 34025716 Free PMC article.
-
Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo.Plant Cell. 2013 Mar;25(3):974-84. doi: 10.1105/tpc.112.108027. Epub 2013 Mar 5. Plant Cell. 2013. PMID: 23463773 Free PMC article.
-
Complete nitrification: insights into the ecophysiology of comammox Nitrospira.Appl Microbiol Biotechnol. 2019 Jan;103(1):177-189. doi: 10.1007/s00253-018-9486-3. Epub 2018 Nov 10. Appl Microbiol Biotechnol. 2019. PMID: 30415428 Free PMC article. Review.
-
Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans.Eukaryot Cell. 2008 Feb;7(2):237-46. doi: 10.1128/EC.00079-07. Epub 2007 Nov 30. Eukaryot Cell. 2008. PMID: 18055915 Free PMC article.
-
Complete genome assembly of Hawai'i environmental nontuberculous mycobacteria reveals unexpected co-isolation with methylobacteria.PLoS One. 2023 Sep 13;18(9):e0291072. doi: 10.1371/journal.pone.0291072. eCollection 2023. PLoS One. 2023. PMID: 37703253 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

