The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization
- PMID: 15563606
- PMCID: PMC545882
- DOI: 10.1091/mbc.e04-07-0553
The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization
Abstract
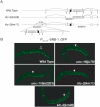
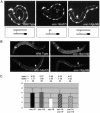
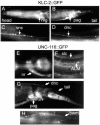
Kinesin-1 is a heterotetramer composed of kinesin heavy chain (KHC) and kinesin light chain (KLC). The Caenorhabditis elegans genome has a single KHC, encoded by the unc-116 gene, and two KLCs, encoded by the klc-1 and klc-2 genes. We show here that UNC-116/KHC and KLC-2 form a complex orthologous to conventional kinesin-1. KLC-2 also binds UNC-16, the C. elegans JIP3/JSAP1 JNK-signaling scaffold protein, and the UNC-14 RUN domain protein. The localization of UNC-16 and UNC-14 depends on kinesin-1 (UNC-116 and KLC-2). Furthermore, mutations in unc-16, klc-2, unc-116, and unc-14 all alter the localization of cargos containing synaptic vesicle markers. Double mutant analysis is consistent with these four genes functioning in the same pathway. Our data support a model whereby UNC-16 and UNC-14 function together as kinesin-1 cargos and regulators for the transport or localization of synaptic vesicle components.
Figures





Similar articles
-
UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans.Neuron. 2001 Dec 6;32(5):787-800. doi: 10.1016/s0896-6273(01)00532-3. Neuron. 2001. PMID: 11738026
-
Regulatory machinery of UNC-33 Ce-CRMP localization in neurites during neuronal development in Caenorhabditis elegans.J Neurochem. 2005 Dec;95(6):1629-41. doi: 10.1111/j.1471-4159.2005.03490.x. Epub 2005 Oct 17. J Neurochem. 2005. PMID: 16236031
-
The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans.Mol Biol Cell. 2004 Aug;15(8):3729-39. doi: 10.1091/mbc.e04-04-0326. Epub 2004 May 21. Mol Biol Cell. 2004. PMID: 15155810 Free PMC article.
-
[MAP kinase cascade regulating synaptic vesicle localization in C. elegans as a model animal].Tanpakushitsu Kakusan Koso. 2002 Sep;47(11):1362-7. Tanpakushitsu Kakusan Koso. 2002. PMID: 12229203 Review. Japanese. No abstract available.
-
The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans.Curr Opin Neurobiol. 2002 Oct;12(5):499-507. doi: 10.1016/s0959-4388(02)00360-4. Curr Opin Neurobiol. 2002. PMID: 12367628 Review.
Cited by
-
Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei.J Cell Biol. 2010 Oct 4;191(1):115-28. doi: 10.1083/jcb.201004118. J Cell Biol. 2010. PMID: 20921138 Free PMC article.
-
Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules.J Biol Chem. 2010 Mar 12;285(11):8155-62. doi: 10.1074/jbc.M109.068247. Epub 2010 Jan 12. J Biol Chem. 2010. PMID: 20071331 Free PMC article.
-
Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain.PLoS One. 2011 May 12;6(5):e19632. doi: 10.1371/journal.pone.0019632. PLoS One. 2011. PMID: 21589871 Free PMC article.
-
The kinesin-3 family motor KLP-4 regulates anterograde trafficking of GLR-1 glutamate receptors in the ventral nerve cord of Caenorhabditis elegans.Mol Biol Cell. 2012 Sep;23(18):3647-62. doi: 10.1091/mbc.E12-04-0334. Epub 2012 Aug 1. Mol Biol Cell. 2012. PMID: 22855524 Free PMC article.
-
MAPK signaling and a mobile scaffold complex regulate AMPA receptor transport to modulate synaptic strength.Cell Rep. 2022 Mar 29;38(13):110577. doi: 10.1016/j.celrep.2022.110577. Cell Rep. 2022. PMID: 35354038 Free PMC article.
References
-
- Akechi, M., Ito, M., Uemura, K., Takamatsu, N., Yamashita, S., Uchiyama, K., Yoshioka, K., and Shiba, T. (2001). Expression of JNK cascade scaffold protein JSAP1 in the mouse nervous system. Neurosci. Res. 39, 391-400. - PubMed
-
- Bowman, A. B., Kamal, A., Ritchings, B. W., Philp, A. V., McGrail, M., Gindhart, J. G., and Goldstein, L.S. (2000). Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103, 583-594. - PubMed
-
- Brady, S. T. (1985). A novel brain ATPase with properties expected for the fast axonal transport motor. Nature 317, 73-75. - PubMed
-
- Byrd, D. T., Kawasaki, M., Walcoff, M., Hisamoto, N., Matsumoto, K., and Jin, Y. (2001). UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32, 787-800. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

